Potassium »
PDB 3m62-3ow2 »
3mz4 »
Potassium in PDB 3mz4: Crystal Structure of D101L MN2+ HDAC8 Complexed with M344
Enzymatic activity of Crystal Structure of D101L MN2+ HDAC8 Complexed with M344
All present enzymatic activity of Crystal Structure of D101L MN2+ HDAC8 Complexed with M344:
3.5.1.98;
3.5.1.98;
Protein crystallography data
The structure of Crystal Structure of D101L MN2+ HDAC8 Complexed with M344, PDB code: 3mz4
was solved by
D.P.Dowling,
S.G.Gattis,
C.A.Fierke,
D.W.Christianson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.35 / 1.84 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 88.353, 91.148, 104.547, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.1 / 24.9 |
Other elements in 3mz4:
The structure of Crystal Structure of D101L MN2+ HDAC8 Complexed with M344 also contains other interesting chemical elements:
| Manganese | (Mn) | 2 atoms |
Potassium Binding Sites:
The binding sites of Potassium atom in the Crystal Structure of D101L MN2+ HDAC8 Complexed with M344
(pdb code 3mz4). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 4 binding sites of Potassium where determined in the Crystal Structure of D101L MN2+ HDAC8 Complexed with M344, PDB code: 3mz4:
Jump to Potassium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Potassium where determined in the Crystal Structure of D101L MN2+ HDAC8 Complexed with M344, PDB code: 3mz4:
Jump to Potassium binding site number: 1; 2; 3; 4;
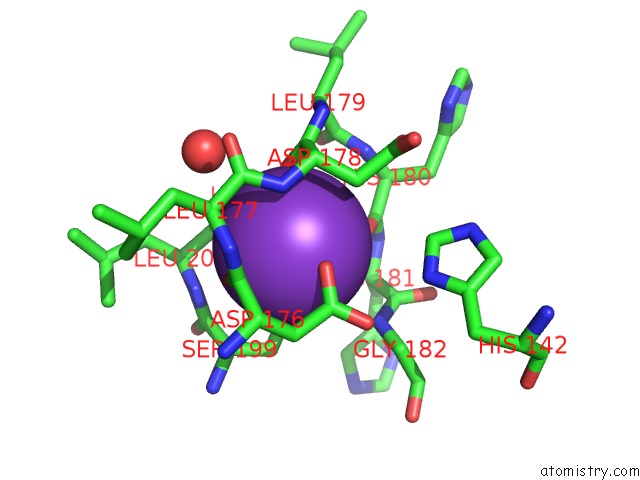
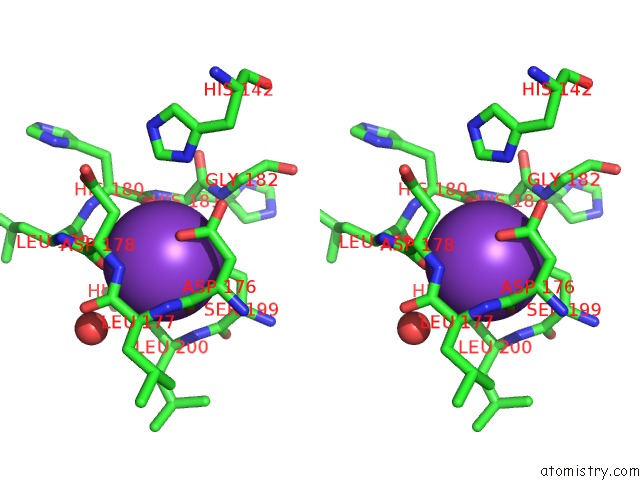
Potassium binding site 1 out of 4 in 3mz4
Go back to
Potassium binding site 1 out
of 4 in the Crystal Structure of D101L MN2+ HDAC8 Complexed with M344
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Crystal Structure of D101L MN2+ HDAC8 Complexed with M344 within 5.0Å range:
|
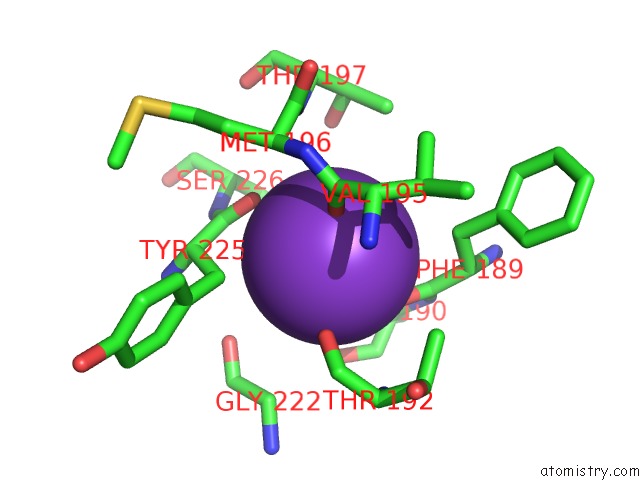
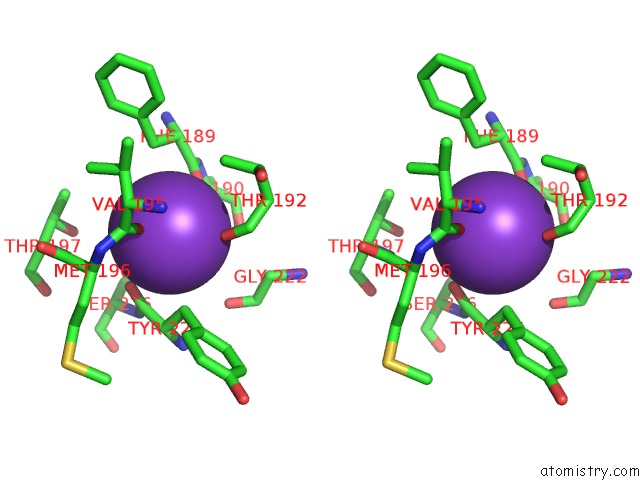
Potassium binding site 2 out of 4 in 3mz4
Go back to
Potassium binding site 2 out
of 4 in the Crystal Structure of D101L MN2+ HDAC8 Complexed with M344
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Crystal Structure of D101L MN2+ HDAC8 Complexed with M344 within 5.0Å range:
|
Potassium binding site 3 out of 4 in 3mz4
Go back to
Potassium binding site 3 out
of 4 in the Crystal Structure of D101L MN2+ HDAC8 Complexed with M344
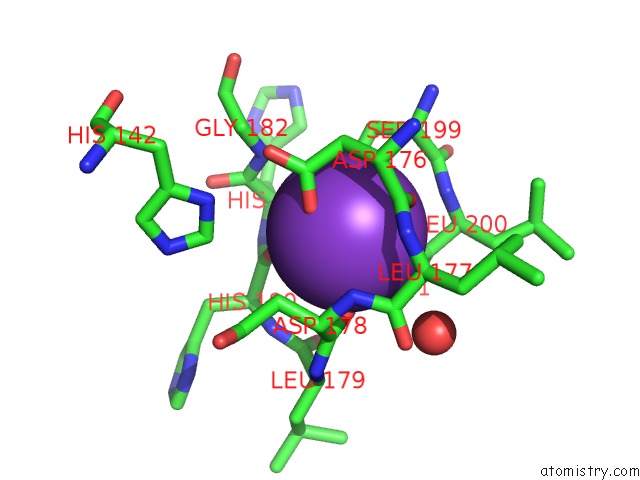
Mono view
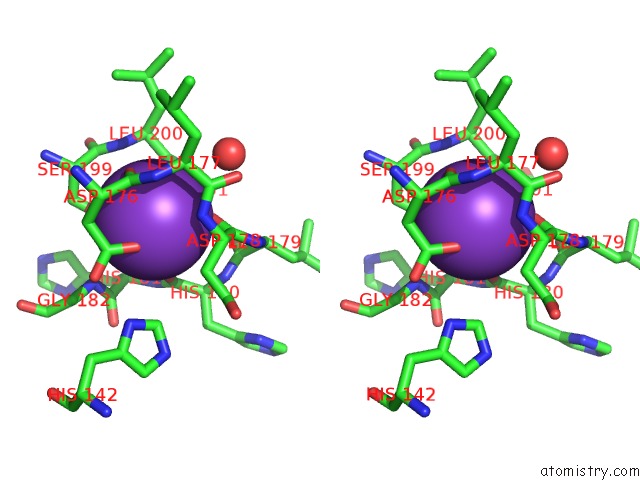
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 3 of Crystal Structure of D101L MN2+ HDAC8 Complexed with M344 within 5.0Å range:
|
Potassium binding site 4 out of 4 in 3mz4
Go back to
Potassium binding site 4 out
of 4 in the Crystal Structure of D101L MN2+ HDAC8 Complexed with M344
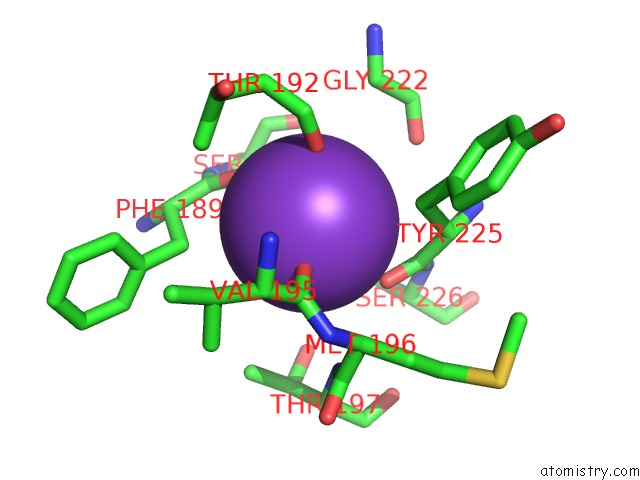
Mono view
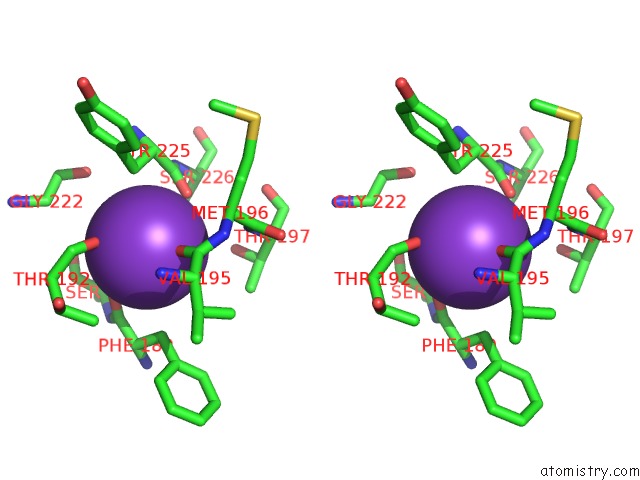
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 4 of Crystal Structure of D101L MN2+ HDAC8 Complexed with M344 within 5.0Å range:
|
Reference:
D.P.Dowling,
S.G.Gattis,
C.A.Fierke,
D.W.Christianson.
Structures of Metal-Substituted Human Histone Deacetylase 8 Provide Mechanistic Inferences on Biological Function. Biochemistry V. 49 5048 2010.
ISSN: ISSN 0006-2960
PubMed: 20545365
DOI: 10.1021/BI1005046
Page generated: Sat Aug 9 05:17:54 2025
ISSN: ISSN 0006-2960
PubMed: 20545365
DOI: 10.1021/BI1005046
Last articles
K in 4LF5K in 4LF4
K in 4LE2
K in 4LCU
K in 4LCA
K in 4LC4
K in 4LBX
K in 4LBG
K in 4LBE
K in 4L6A