Potassium »
PDB 6n93-6pc3 »
6oxd »
Potassium in PDB 6oxd: Structure of Mycobacterium Tuberculosis Methylmalonyl-Coa Mutase with Adenosyl Cobalamin
Enzymatic activity of Structure of Mycobacterium Tuberculosis Methylmalonyl-Coa Mutase with Adenosyl Cobalamin
All present enzymatic activity of Structure of Mycobacterium Tuberculosis Methylmalonyl-Coa Mutase with Adenosyl Cobalamin:
5.4.99.2;
5.4.99.2;
Protein crystallography data
The structure of Structure of Mycobacterium Tuberculosis Methylmalonyl-Coa Mutase with Adenosyl Cobalamin, PDB code: 6oxd
was solved by
M.Purchal,
M.Ruetz,
R.Banerjee,
M.Koutmos,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.80 / 2.00 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 76.576, 104.957, 194.088, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.9 / 19.7 |
Other elements in 6oxd:
The structure of Structure of Mycobacterium Tuberculosis Methylmalonyl-Coa Mutase with Adenosyl Cobalamin also contains other interesting chemical elements:
| Cobalt | (Co) | 1 atom |
Potassium Binding Sites:
The binding sites of Potassium atom in the Structure of Mycobacterium Tuberculosis Methylmalonyl-Coa Mutase with Adenosyl Cobalamin
(pdb code 6oxd). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total only one binding site of Potassium was determined in the Structure of Mycobacterium Tuberculosis Methylmalonyl-Coa Mutase with Adenosyl Cobalamin, PDB code: 6oxd:
In total only one binding site of Potassium was determined in the Structure of Mycobacterium Tuberculosis Methylmalonyl-Coa Mutase with Adenosyl Cobalamin, PDB code: 6oxd:
Potassium binding site 1 out of 1 in 6oxd
Go back to
Potassium binding site 1 out
of 1 in the Structure of Mycobacterium Tuberculosis Methylmalonyl-Coa Mutase with Adenosyl Cobalamin
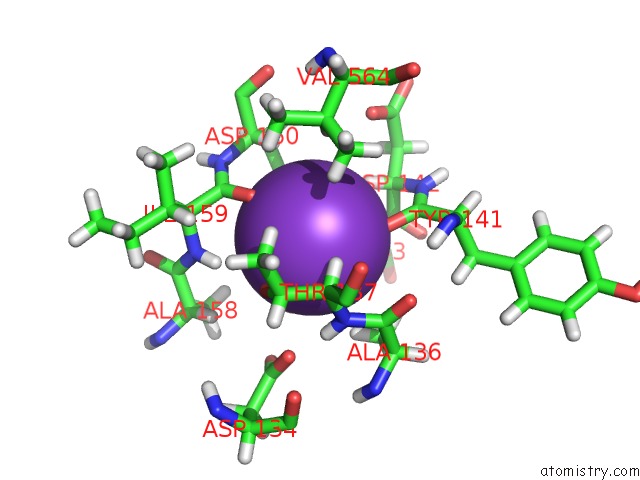
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Structure of Mycobacterium Tuberculosis Methylmalonyl-Coa Mutase with Adenosyl Cobalamin within 5.0Å range:
|
Reference:
M.Ruetz,
G.C.Campanello,
M.Purchal,
H.Shen,
L.Mcdevitt,
H.Gouda,
S.Wakabayashi,
J.Zhu,
E.J.Rubin,
K.Warncke,
V.K.Mootha,
M.Koutmos,
R.Banerjee.
Itaconyl-Coa Forms A Stable Biradical in Methylmalonyl-Coa Mutase and Derails Its Activity and Repair. Science V. 366 589 2019.
ISSN: ESSN 1095-9203
PubMed: 31672889
DOI: 10.1126/SCIENCE.AAY0934
Page generated: Sat Aug 9 11:44:43 2025
ISSN: ESSN 1095-9203
PubMed: 31672889
DOI: 10.1126/SCIENCE.AAY0934
Last articles
Mg in 1MQ4Mg in 1MPO
Mg in 1MQ3
Mg in 1MMS
Mg in 1MPN
Mg in 1MOW
Mg in 1MPM
Mg in 1MOJ
Mg in 1MOG
Mg in 1MNZ