Potassium »
PDB 6n93-6pc3 »
6nzg »
Potassium in PDB 6nzg: Bacteroides Uniformis Beta-Glucuronidase 2 Covalently Bound to Cyclophellitol-6-Carboxylate Aziridine
Enzymatic activity of Bacteroides Uniformis Beta-Glucuronidase 2 Covalently Bound to Cyclophellitol-6-Carboxylate Aziridine
All present enzymatic activity of Bacteroides Uniformis Beta-Glucuronidase 2 Covalently Bound to Cyclophellitol-6-Carboxylate Aziridine:
3.2.1.23; 3.2.1.31;
3.2.1.23; 3.2.1.31;
Protein crystallography data
The structure of Bacteroides Uniformis Beta-Glucuronidase 2 Covalently Bound to Cyclophellitol-6-Carboxylate Aziridine, PDB code: 6nzg
was solved by
S.J.Pellock,
P.B.Jariwala,
M.R.Redinbo,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.46 / 2.43 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 74.536, 142.407, 180.664, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17 / 22.1 |
Other elements in 6nzg:
The structure of Bacteroides Uniformis Beta-Glucuronidase 2 Covalently Bound to Cyclophellitol-6-Carboxylate Aziridine also contains other interesting chemical elements:
| Calcium | (Ca) | 2 atoms |
Potassium Binding Sites:
The binding sites of Potassium atom in the Bacteroides Uniformis Beta-Glucuronidase 2 Covalently Bound to Cyclophellitol-6-Carboxylate Aziridine
(pdb code 6nzg). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 2 binding sites of Potassium where determined in the Bacteroides Uniformis Beta-Glucuronidase 2 Covalently Bound to Cyclophellitol-6-Carboxylate Aziridine, PDB code: 6nzg:
Jump to Potassium binding site number: 1; 2;
In total 2 binding sites of Potassium where determined in the Bacteroides Uniformis Beta-Glucuronidase 2 Covalently Bound to Cyclophellitol-6-Carboxylate Aziridine, PDB code: 6nzg:
Jump to Potassium binding site number: 1; 2;
Potassium binding site 1 out of 2 in 6nzg
Go back to
Potassium binding site 1 out
of 2 in the Bacteroides Uniformis Beta-Glucuronidase 2 Covalently Bound to Cyclophellitol-6-Carboxylate Aziridine

Mono view
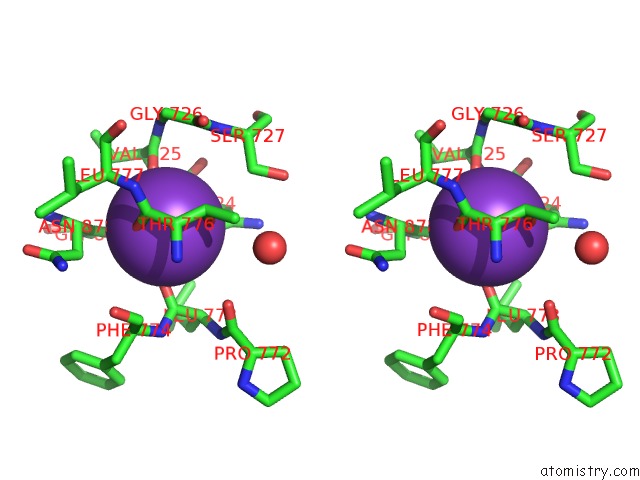
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Bacteroides Uniformis Beta-Glucuronidase 2 Covalently Bound to Cyclophellitol-6-Carboxylate Aziridine within 5.0Å range:
|
Potassium binding site 2 out of 2 in 6nzg
Go back to
Potassium binding site 2 out
of 2 in the Bacteroides Uniformis Beta-Glucuronidase 2 Covalently Bound to Cyclophellitol-6-Carboxylate Aziridine

Mono view
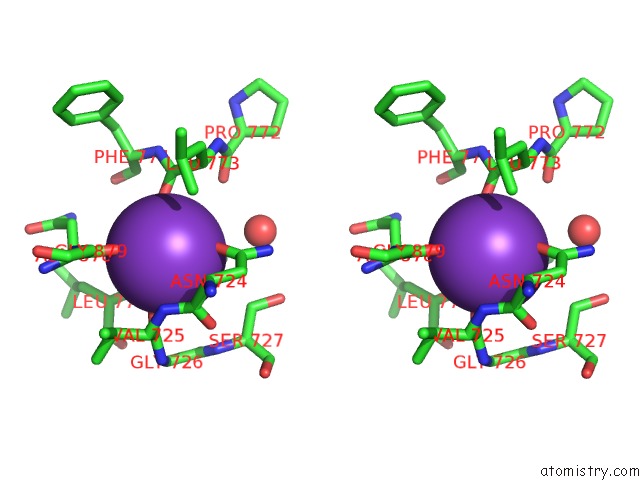
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Bacteroides Uniformis Beta-Glucuronidase 2 Covalently Bound to Cyclophellitol-6-Carboxylate Aziridine within 5.0Å range:
|
Reference:
P.B.Jariwala,
S.J.Pellock,
D.Goldfarb,
E.W.Cloer,
M.Artola,
J.B.Simpson,
A.P.Bhatt,
W.G.Walton,
L.R.Roberts,
M.B.Major,
G.J.Davies,
H.S.Overkleeft,
M.R.Redinbo.
Discovering the Microbial Enzymes Driving Drug Toxicity with Activity-Based Protein Profiling. Acs Chem.Biol. 2019.
ISSN: ESSN 1554-8937
PubMed: 31774274
DOI: 10.1021/ACSCHEMBIO.9B00788
Page generated: Sat Aug 9 11:41:15 2025
ISSN: ESSN 1554-8937
PubMed: 31774274
DOI: 10.1021/ACSCHEMBIO.9B00788
Last articles
K in 9FYEK in 9FT7
K in 9FQ1
K in 9FM9
K in 9EX3
K in 9F90
K in 9ES6
K in 9EWD
K in 9ETN
K in 9ESI