Potassium »
PDB 1gup-1k4c »
1guq »
Potassium in PDB 1guq: Structure of Nucleotidyltransferase Complexed with Udp-Glucose
Enzymatic activity of Structure of Nucleotidyltransferase Complexed with Udp-Glucose
All present enzymatic activity of Structure of Nucleotidyltransferase Complexed with Udp-Glucose:
2.7.7.10;
2.7.7.10;
Protein crystallography data
The structure of Structure of Nucleotidyltransferase Complexed with Udp-Glucose, PDB code: 1guq
was solved by
J.B.Thoden,
I.Rayment,
H.Holden,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 1.80 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 68.400, 57.500, 188.900, 90.00, 100.13, 90.00 |
| R / Rfree (%) | n/a / n/a |
Other elements in 1guq:
The structure of Structure of Nucleotidyltransferase Complexed with Udp-Glucose also contains other interesting chemical elements:
| Iron | (Fe) | 4 atoms |
| Zinc | (Zn) | 4 atoms |
Potassium Binding Sites:
The binding sites of Potassium atom in the Structure of Nucleotidyltransferase Complexed with Udp-Glucose
(pdb code 1guq). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 4 binding sites of Potassium where determined in the Structure of Nucleotidyltransferase Complexed with Udp-Glucose, PDB code: 1guq:
Jump to Potassium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Potassium where determined in the Structure of Nucleotidyltransferase Complexed with Udp-Glucose, PDB code: 1guq:
Jump to Potassium binding site number: 1; 2; 3; 4;
Potassium binding site 1 out of 4 in 1guq
Go back to
Potassium binding site 1 out
of 4 in the Structure of Nucleotidyltransferase Complexed with Udp-Glucose
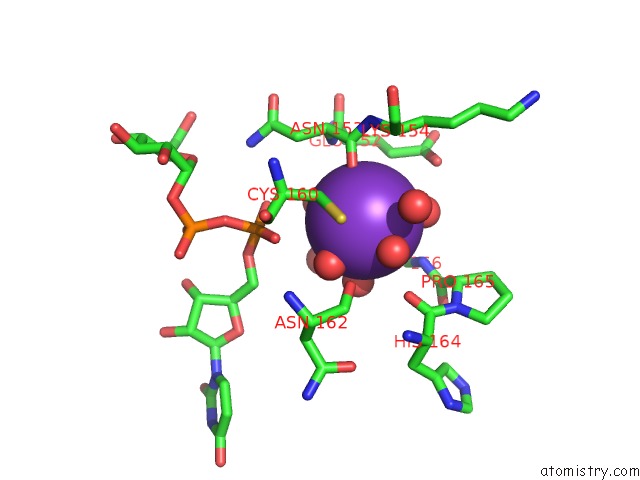
Mono view
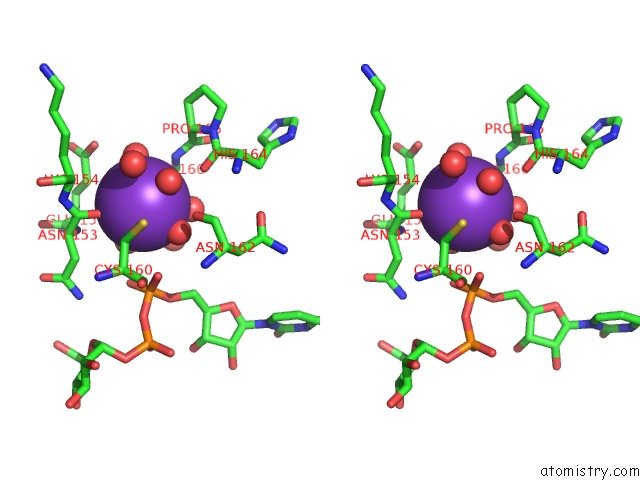
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Structure of Nucleotidyltransferase Complexed with Udp-Glucose within 5.0Å range:
|
Potassium binding site 2 out of 4 in 1guq
Go back to
Potassium binding site 2 out
of 4 in the Structure of Nucleotidyltransferase Complexed with Udp-Glucose
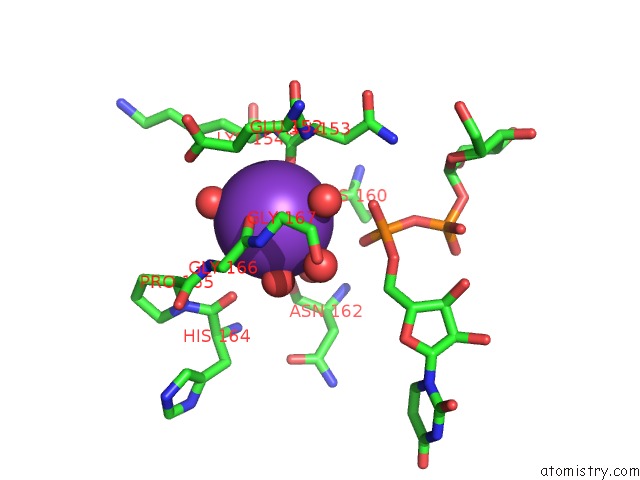
Mono view
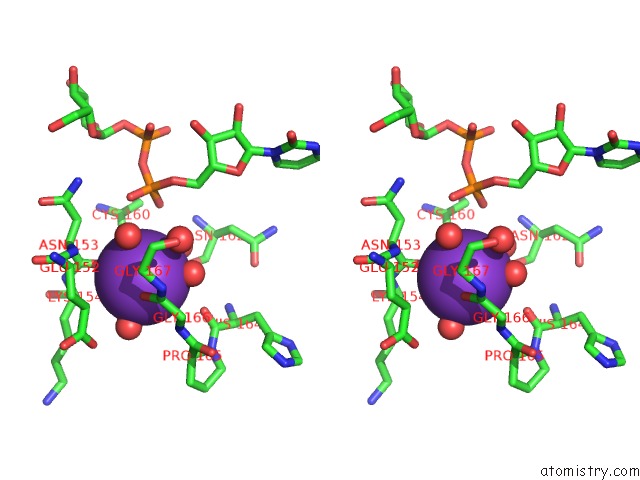
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Structure of Nucleotidyltransferase Complexed with Udp-Glucose within 5.0Å range:
|
Potassium binding site 3 out of 4 in 1guq
Go back to
Potassium binding site 3 out
of 4 in the Structure of Nucleotidyltransferase Complexed with Udp-Glucose
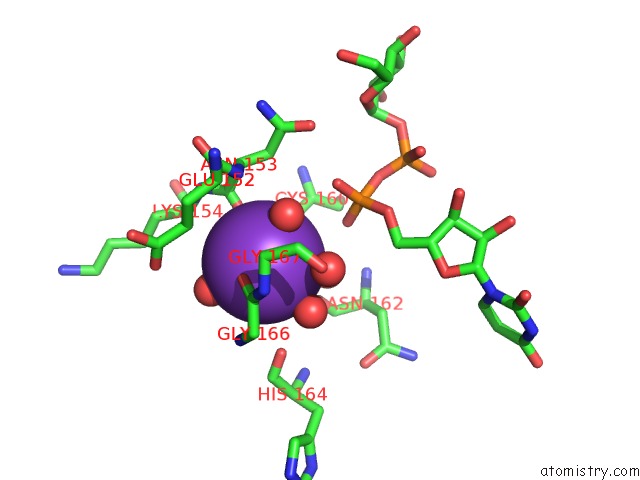
Mono view
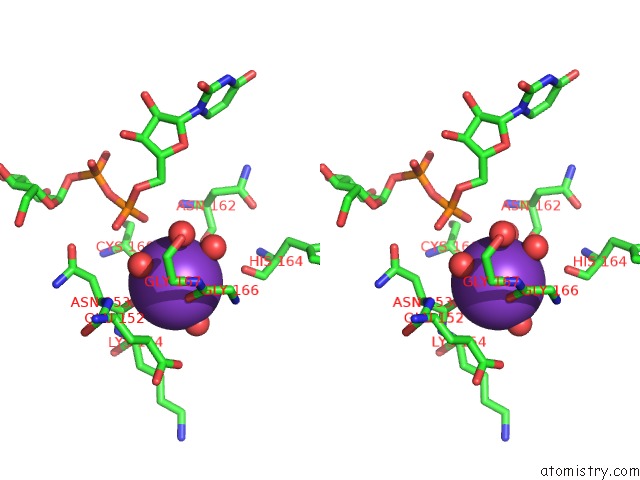
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 3 of Structure of Nucleotidyltransferase Complexed with Udp-Glucose within 5.0Å range:
|
Potassium binding site 4 out of 4 in 1guq
Go back to
Potassium binding site 4 out
of 4 in the Structure of Nucleotidyltransferase Complexed with Udp-Glucose
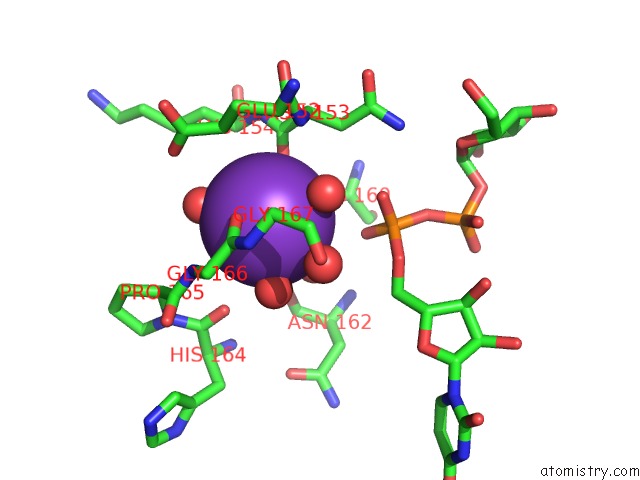
Mono view
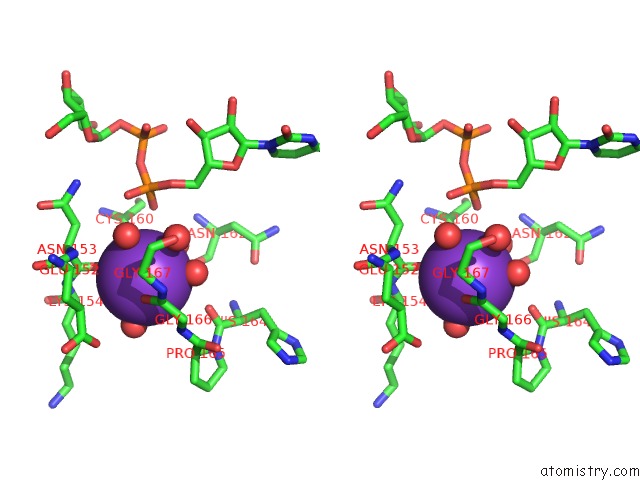
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 4 of Structure of Nucleotidyltransferase Complexed with Udp-Glucose within 5.0Å range:
|
Reference:
J.B.Thoden,
F.J.Ruzicka,
P.A.Frey,
I.Rayment,
H.M.Holden.
Structural Analysis of the H166G Site-Directed Mutant of Galactose-1-Phosphate Uridylyltransferase Complexed with Either Udp-Glucose or Udp-Galactose: Detailed Description of the Nucleotide Sugar Binding Site. Biochemistry V. 36 1212 1997.
ISSN: ISSN 0006-2960
PubMed: 9063869
DOI: 10.1021/BI9626517
Page generated: Sat Aug 9 01:56:25 2025
ISSN: ISSN 0006-2960
PubMed: 9063869
DOI: 10.1021/BI9626517
Last articles
K in 4BYZK in 4BYG
K in 4BVA
K in 4BGA
K in 4BV8
K in 4BKX
K in 4BGB
K in 4BI3
K in 4BH5
K in 4B3T