Potassium »
PDB 7xxk-7zgo »
7z4i »
Potassium in PDB 7z4i: SPCAS9 Bound to 16-Nucleotide Complementary Dna Substrate
Potassium Binding Sites:
The binding sites of Potassium atom in the SPCAS9 Bound to 16-Nucleotide Complementary Dna Substrate
(pdb code 7z4i). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 2 binding sites of Potassium where determined in the SPCAS9 Bound to 16-Nucleotide Complementary Dna Substrate, PDB code: 7z4i:
Jump to Potassium binding site number: 1; 2;
In total 2 binding sites of Potassium where determined in the SPCAS9 Bound to 16-Nucleotide Complementary Dna Substrate, PDB code: 7z4i:
Jump to Potassium binding site number: 1; 2;
Potassium binding site 1 out of 2 in 7z4i
Go back to
Potassium binding site 1 out
of 2 in the SPCAS9 Bound to 16-Nucleotide Complementary Dna Substrate
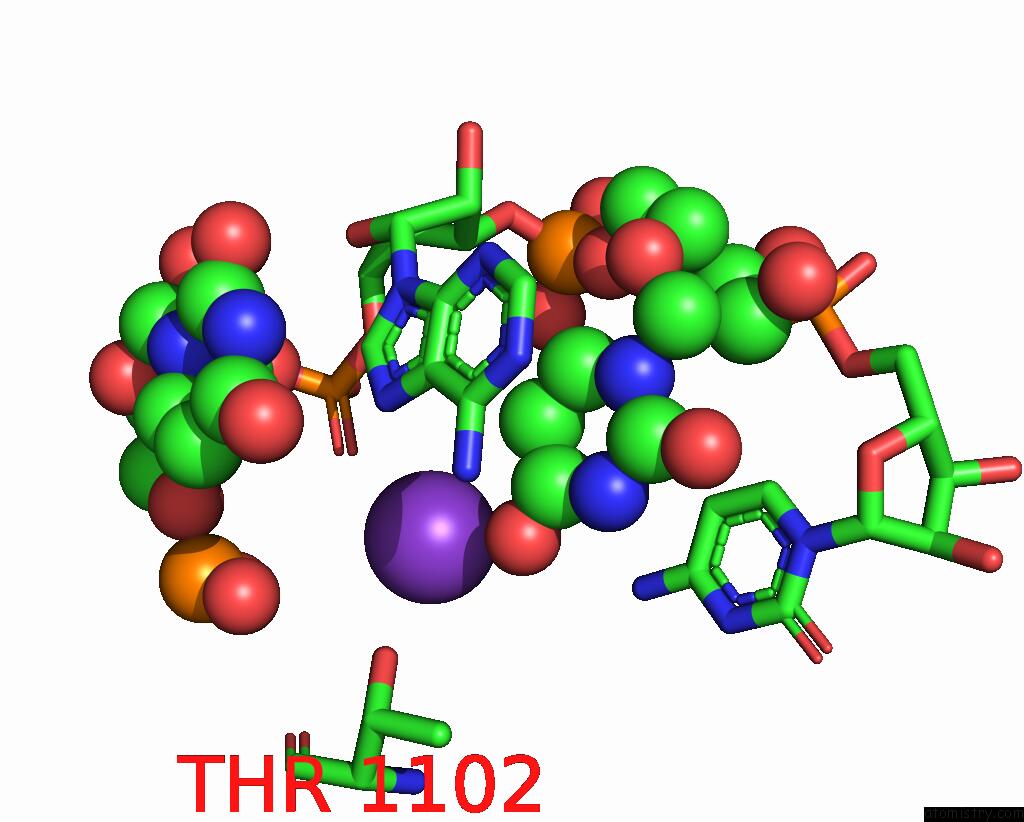
Mono view
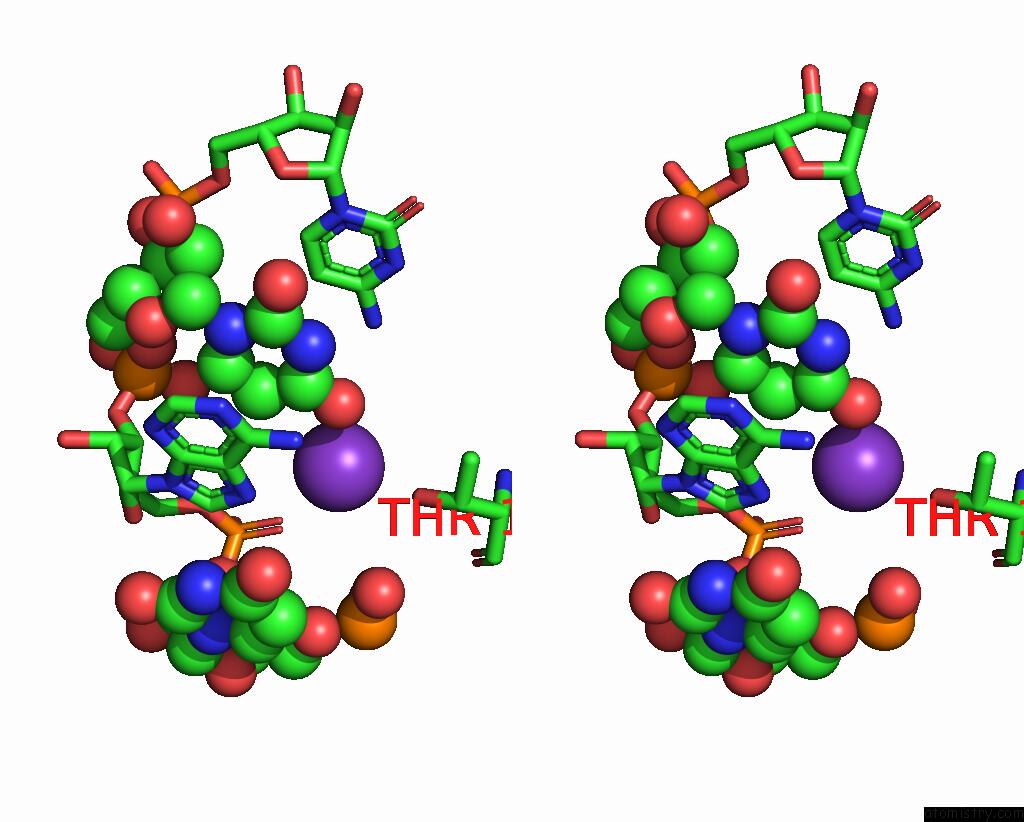
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of SPCAS9 Bound to 16-Nucleotide Complementary Dna Substrate within 5.0Å range:
|
Potassium binding site 2 out of 2 in 7z4i
Go back to
Potassium binding site 2 out
of 2 in the SPCAS9 Bound to 16-Nucleotide Complementary Dna Substrate
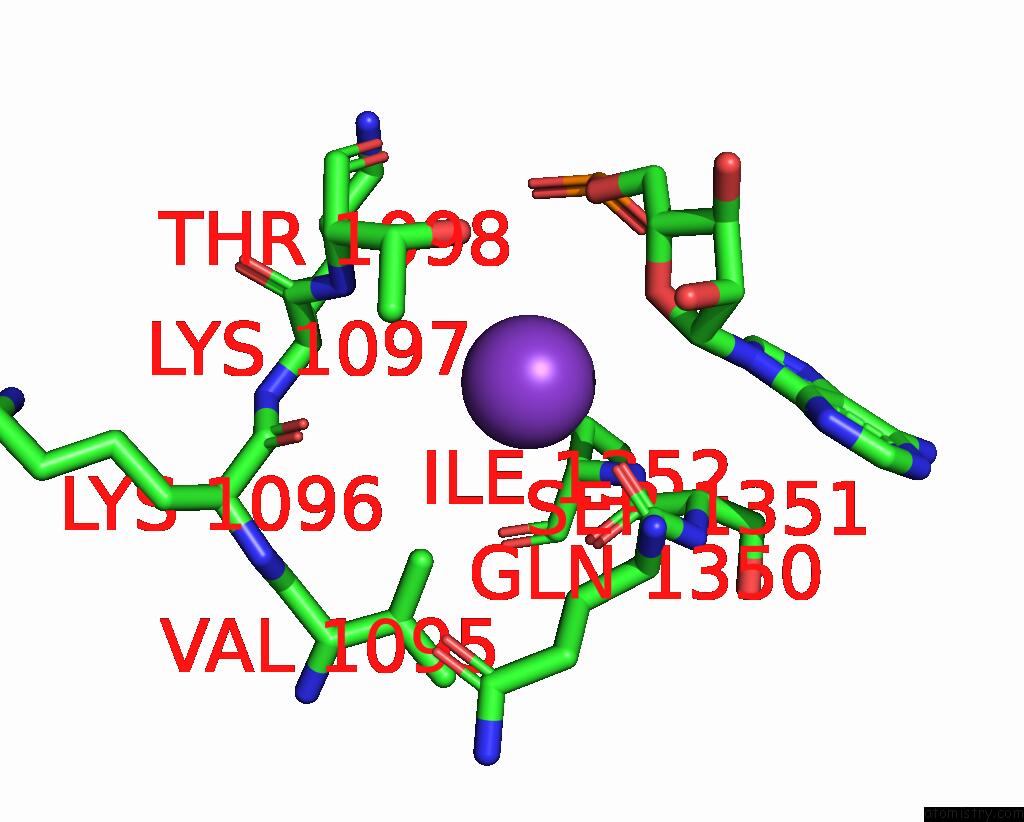
Mono view
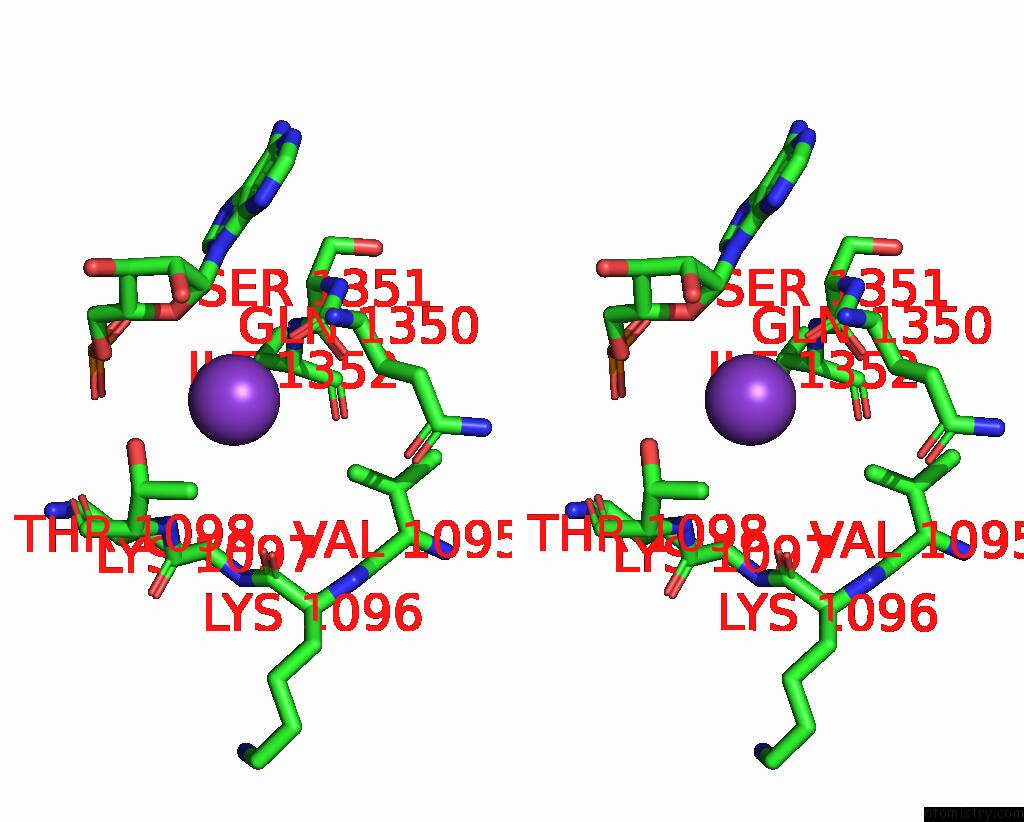
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of SPCAS9 Bound to 16-Nucleotide Complementary Dna Substrate within 5.0Å range:
|
Reference:
M.Pacesa,
L.Loeff,
I.Querques,
L.M.Muckenfuss,
M.Sawicka,
M.Jinek.
R-Loop Formation and Conformational Activation Mechanisms of CAS9. Nature V. 609 191 2022.
ISSN: ESSN 1476-4687
PubMed: 36002571
DOI: 10.1038/S41586-022-05114-0
Page generated: Sat Aug 9 15:22:00 2025
ISSN: ESSN 1476-4687
PubMed: 36002571
DOI: 10.1038/S41586-022-05114-0
Last articles
Mg in 6C5NMg in 6C55
Mg in 6C2W
Mg in 6C3P
Mg in 6C3O
Mg in 6C2C
Mg in 6C2X
Mg in 6C25
Mg in 6C1X
Mg in 6C1H