Potassium »
PDB 7qr3-7rut »
7qsm »
Potassium in PDB 7qsm: Bovine Complex I in Lipid Nanodisc, Deactive-Ligand (Composite)
Enzymatic activity of Bovine Complex I in Lipid Nanodisc, Deactive-Ligand (Composite)
All present enzymatic activity of Bovine Complex I in Lipid Nanodisc, Deactive-Ligand (Composite):
1.6.5.3; 1.6.99.3; 7.1.1.2;
1.6.5.3; 1.6.99.3; 7.1.1.2;
Other elements in 7qsm:
The structure of Bovine Complex I in Lipid Nanodisc, Deactive-Ligand (Composite) also contains other interesting chemical elements:
| Iron | (Fe) | 28 atoms |
| Zinc | (Zn) | 1 atom |
| Magnesium | (Mg) | 1 atom |
Potassium Binding Sites:
The binding sites of Potassium atom in the Bovine Complex I in Lipid Nanodisc, Deactive-Ligand (Composite)
(pdb code 7qsm). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total only one binding site of Potassium was determined in the Bovine Complex I in Lipid Nanodisc, Deactive-Ligand (Composite), PDB code: 7qsm:
In total only one binding site of Potassium was determined in the Bovine Complex I in Lipid Nanodisc, Deactive-Ligand (Composite), PDB code: 7qsm:
Potassium binding site 1 out of 1 in 7qsm
Go back to
Potassium binding site 1 out
of 1 in the Bovine Complex I in Lipid Nanodisc, Deactive-Ligand (Composite)

Mono view
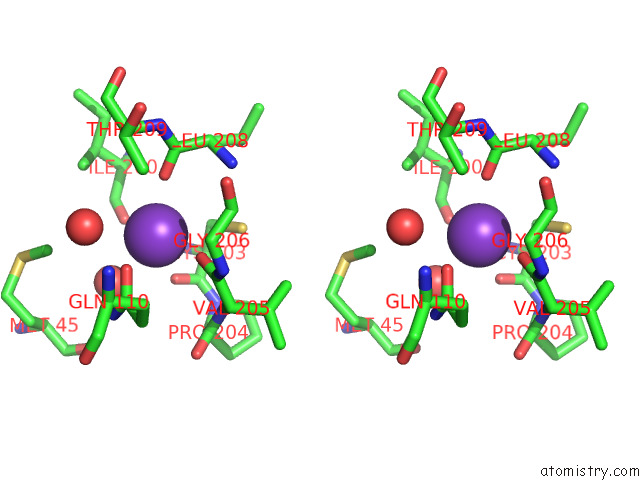
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Bovine Complex I in Lipid Nanodisc, Deactive-Ligand (Composite) within 5.0Å range:
|
Reference:
I.Chung,
J.J.Wright,
H.R.Bridges,
B.S.Ivanov,
O.Biner,
C.S.Pereira,
G.M.Arantes,
J.Hirst.
Cryo-Em Structures Define Ubiquinone-10 Binding to Mitochondrial Complex I and Conformational Transitions Accompanying Q-Site Occupancy. Nat Commun V. 13 2758 2022.
ISSN: ESSN 2041-1723
PubMed: 35589726
DOI: 10.1038/S41467-022-30506-1
Page generated: Sat Aug 9 14:39:27 2025
ISSN: ESSN 2041-1723
PubMed: 35589726
DOI: 10.1038/S41467-022-30506-1
Last articles
Mg in 5HXTMg in 5HX1
Mg in 5HXP
Mg in 5HVK
Mg in 5HWE
Mg in 5HV3
Mg in 5HV1
Mg in 5HUH
Mg in 5HTK
Mg in 5HTO