Potassium »
PDB 5x24-5ze2 »
5xrz »
Potassium in PDB 5xrz: Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52
Protein crystallography data
The structure of Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52, PDB code: 5xrz
was solved by
M.Saotome,
K.Saito,
T.Yasuda,
S.Sugiyama,
H.Kurumizaka,
W.Kagawa,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.70 / 3.60 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 67.621, 101.420, 101.710, 91.89, 108.68, 108.17 |
| R / Rfree (%) | 21.7 / 25.7 |
Potassium Binding Sites:
The binding sites of Potassium atom in the Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52
(pdb code 5xrz). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 10 binding sites of Potassium where determined in the Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52, PDB code: 5xrz:
Jump to Potassium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
In total 10 binding sites of Potassium where determined in the Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52, PDB code: 5xrz:
Jump to Potassium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
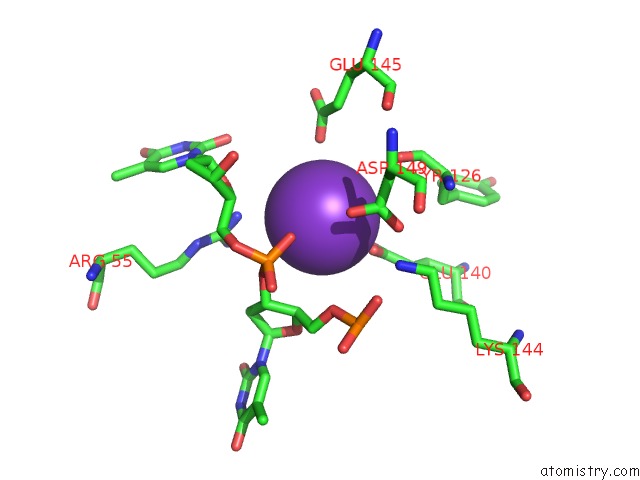
Potassium binding site 1 out of 10 in 5xrz
Go back to
Potassium binding site 1 out
of 10 in the Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52 within 5.0Å range:
|
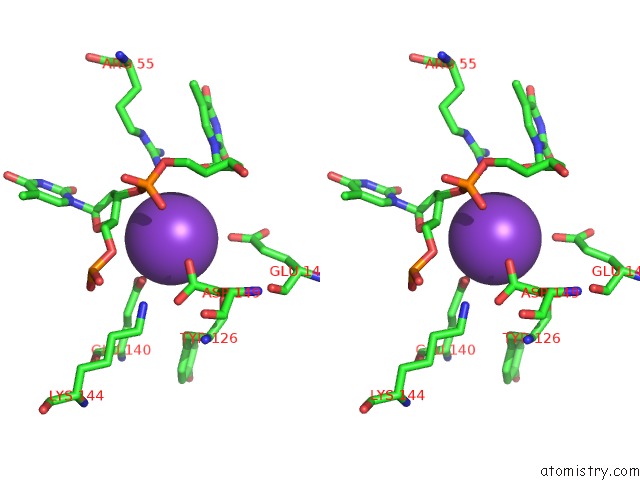
Potassium binding site 2 out of 10 in 5xrz
Go back to
Potassium binding site 2 out
of 10 in the Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52 within 5.0Å range:
|
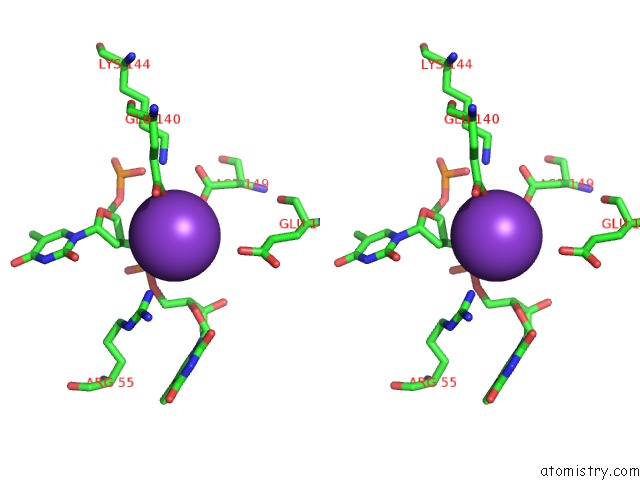
Potassium binding site 3 out of 10 in 5xrz
Go back to
Potassium binding site 3 out
of 10 in the Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 3 of Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52 within 5.0Å range:
|
Potassium binding site 4 out of 10 in 5xrz
Go back to
Potassium binding site 4 out
of 10 in the Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 4 of Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52 within 5.0Å range:
|
Potassium binding site 5 out of 10 in 5xrz
Go back to
Potassium binding site 5 out
of 10 in the Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 5 of Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52 within 5.0Å range:
|
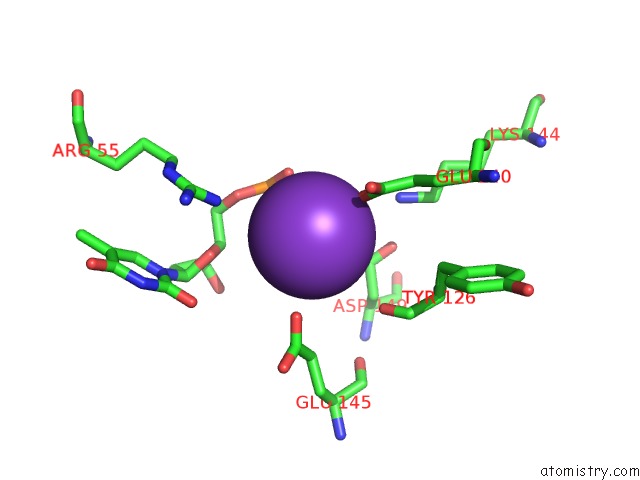
Potassium binding site 6 out of 10 in 5xrz
Go back to
Potassium binding site 6 out
of 10 in the Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52
Mono view
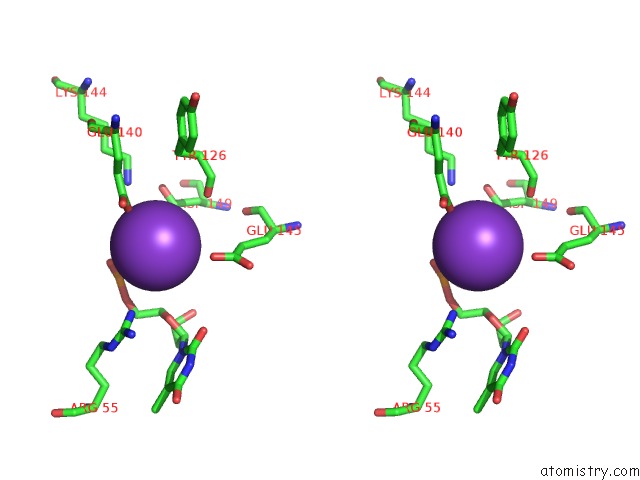
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 6 of Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52 within 5.0Å range:
|
Potassium binding site 7 out of 10 in 5xrz
Go back to
Potassium binding site 7 out
of 10 in the Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52
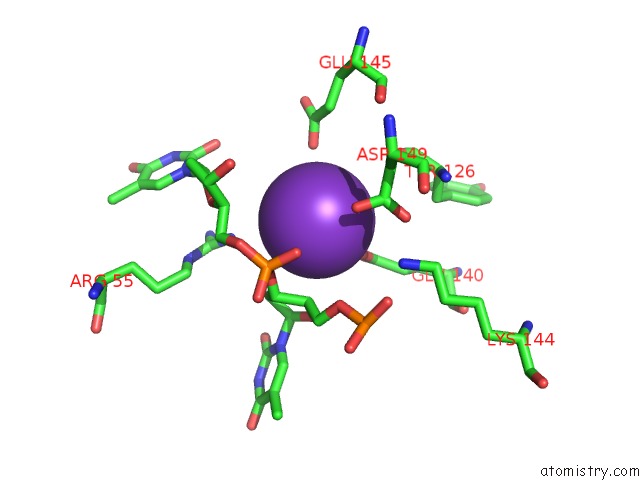
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 7 of Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52 within 5.0Å range:
|
Potassium binding site 8 out of 10 in 5xrz
Go back to
Potassium binding site 8 out
of 10 in the Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52

Mono view
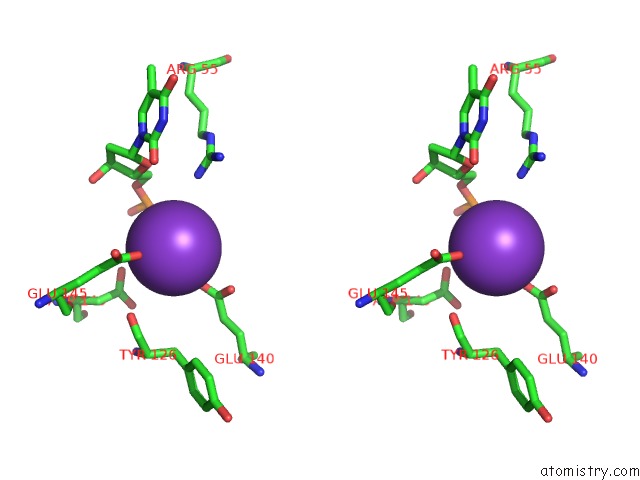
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 8 of Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52 within 5.0Å range:
|
Potassium binding site 9 out of 10 in 5xrz
Go back to
Potassium binding site 9 out
of 10 in the Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52
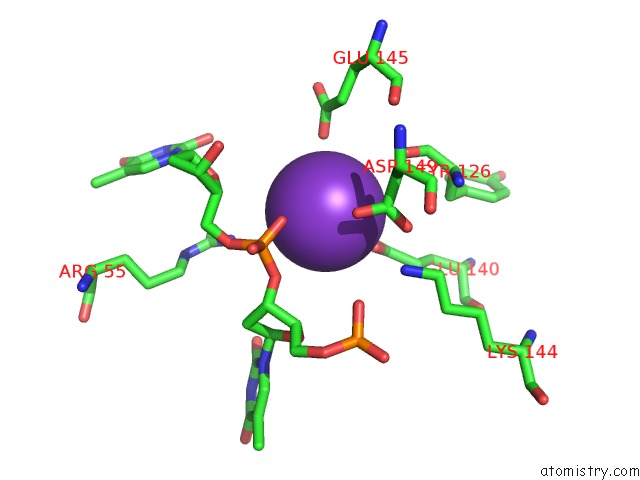
Mono view
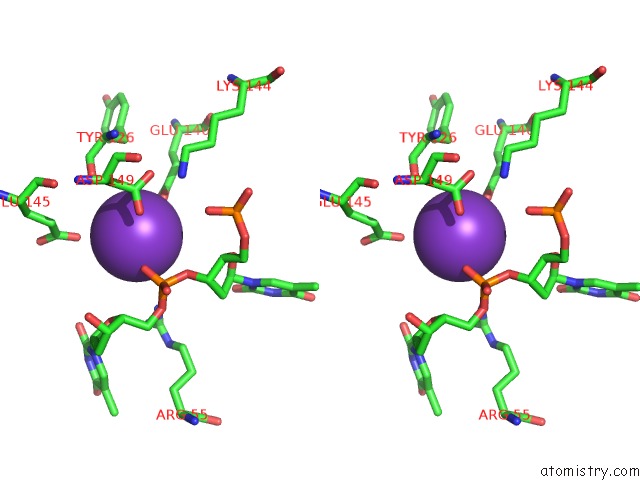
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 9 of Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52 within 5.0Å range:
|
Potassium binding site 10 out of 10 in 5xrz
Go back to
Potassium binding site 10 out
of 10 in the Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52

Mono view
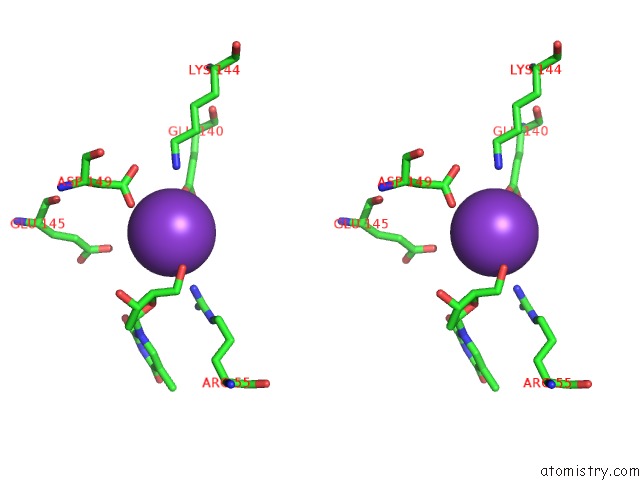
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 10 of Structure of A Ssdna Bound to the Inner Dna Binding Site of RAD52 within 5.0Å range:
|
Reference:
M.Saotome,
K.Saito,
T.Yasuda,
H.Ohtomo,
S.Sugiyama,
Y.Nishimura,
H.Kurumizaka,
W.Kagawa.
Structural Basis of Homology-Directed Dna Repair Mediated By RAD52 Iscience V. 3 50 2018.
ISSN: ESSN 2589-0042
PubMed: 30428330
DOI: 10.1016/J.ISCI.2018.04.005
Page generated: Sat Aug 9 10:14:25 2025
ISSN: ESSN 2589-0042
PubMed: 30428330
DOI: 10.1016/J.ISCI.2018.04.005
Last articles
Zn in 2YBGZn in 2YBP
Zn in 2YBK
Zn in 2YAV
Zn in 2Y7I
Zn in 2YBB
Zn in 2YB5
Zn in 2YB9
Zn in 2YAC
Zn in 2Y8B