Potassium »
PDB 5dkp-5f0g »
5eek »
Potassium in PDB 5eek: Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with Trichostatin A
Enzymatic activity of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with Trichostatin A
All present enzymatic activity of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with Trichostatin A:
3.5.1.98;
3.5.1.98;
Protein crystallography data
The structure of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with Trichostatin A, PDB code: 5eek
was solved by
Y.Hai,
D.W.Christianson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 44.01 / 1.59 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 83.881, 94.429, 51.698, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 13 / 16.3 |
Other elements in 5eek:
The structure of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with Trichostatin A also contains other interesting chemical elements:
| Zinc | (Zn) | 1 atom |
| Iodine | (I) | 31 atoms |
| Chlorine | (Cl) | 1 atom |
Potassium Binding Sites:
The binding sites of Potassium atom in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with Trichostatin A
(pdb code 5eek). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 2 binding sites of Potassium where determined in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with Trichostatin A, PDB code: 5eek:
Jump to Potassium binding site number: 1; 2;
In total 2 binding sites of Potassium where determined in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with Trichostatin A, PDB code: 5eek:
Jump to Potassium binding site number: 1; 2;
Potassium binding site 1 out of 2 in 5eek
Go back to
Potassium binding site 1 out
of 2 in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with Trichostatin A
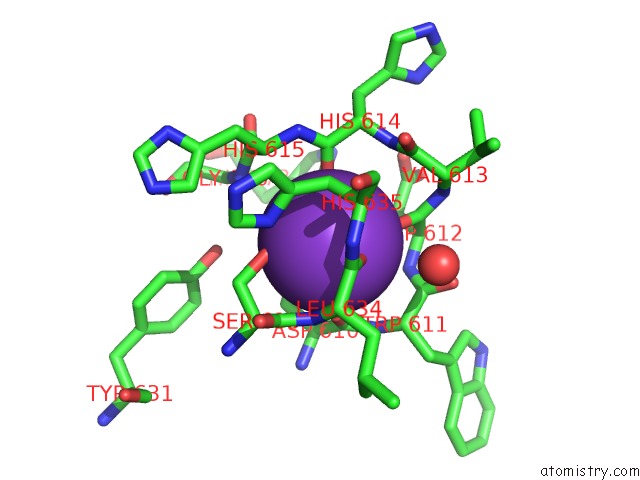
Mono view
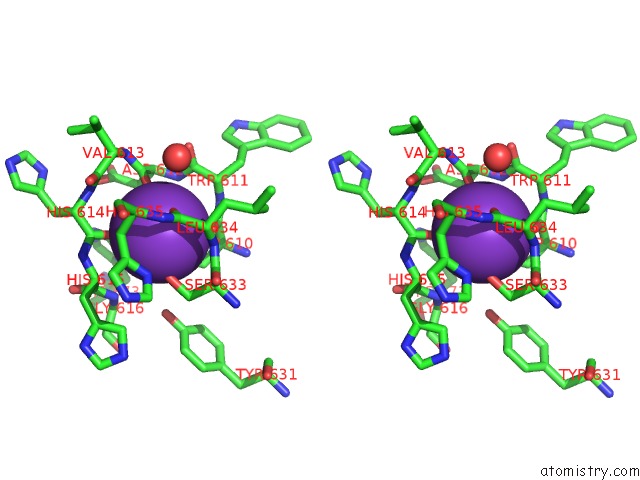
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with Trichostatin A within 5.0Å range:
|
Potassium binding site 2 out of 2 in 5eek
Go back to
Potassium binding site 2 out
of 2 in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with Trichostatin A
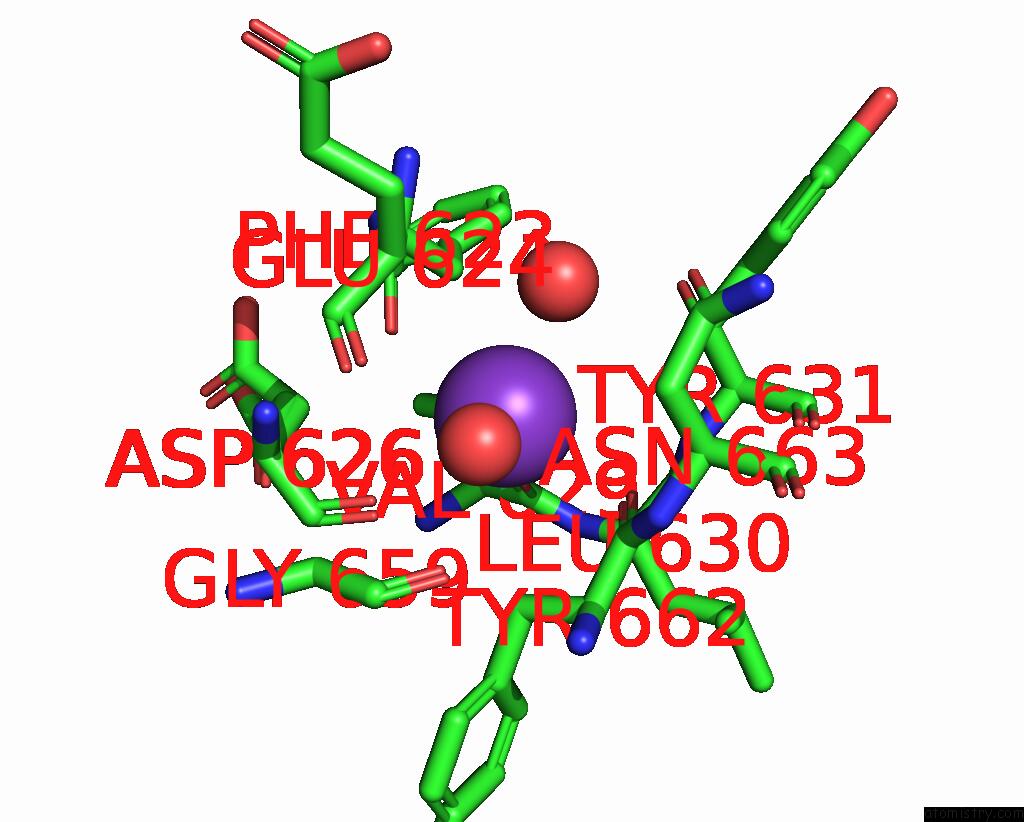
Mono view
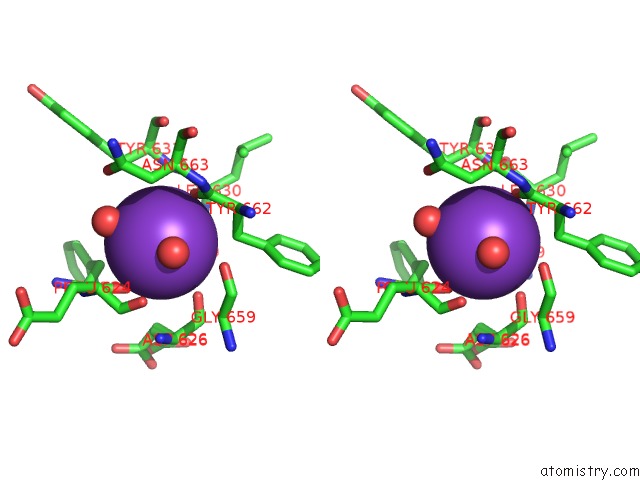
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with Trichostatin A within 5.0Å range:
|
Reference:
Y.Hai,
D.W.Christianson.
Histone Deacetylase 6 Structure and Molecular Basis of Catalysis and Inhibition. Nat.Chem.Biol. V. 12 741 2016.
ISSN: ESSN 1552-4469
PubMed: 27454933
DOI: 10.1038/NCHEMBIO.2134
Page generated: Sat Aug 9 08:53:55 2025
ISSN: ESSN 1552-4469
PubMed: 27454933
DOI: 10.1038/NCHEMBIO.2134
Last articles
K in 6P9VK in 6P0Y
K in 6OZI
K in 6P1M
K in 6OZJ
K in 6OZR
K in 6OXD
K in 6OF8
K in 6OSR
K in 6OVL