Potassium »
PDB 4k93-4l4c »
4kax »
Potassium in PDB 4kax: Crystal Structure of the GRP1 pH Domain in Complex with ARF6-Gtp
Protein crystallography data
The structure of Crystal Structure of the GRP1 pH Domain in Complex with ARF6-Gtp, PDB code: 4kax
was solved by
D.G.Lambright,
A.W.Malaby,
B.Van Den Berg,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 1.85 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 56.595, 56.595, 274.371, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20 / 23.5 |
Other elements in 4kax:
The structure of Crystal Structure of the GRP1 pH Domain in Complex with ARF6-Gtp also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Potassium Binding Sites:
The binding sites of Potassium atom in the Crystal Structure of the GRP1 pH Domain in Complex with ARF6-Gtp
(pdb code 4kax). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total only one binding site of Potassium was determined in the Crystal Structure of the GRP1 pH Domain in Complex with ARF6-Gtp, PDB code: 4kax:
In total only one binding site of Potassium was determined in the Crystal Structure of the GRP1 pH Domain in Complex with ARF6-Gtp, PDB code: 4kax:
Potassium binding site 1 out of 1 in 4kax
Go back to
Potassium binding site 1 out
of 1 in the Crystal Structure of the GRP1 pH Domain in Complex with ARF6-Gtp
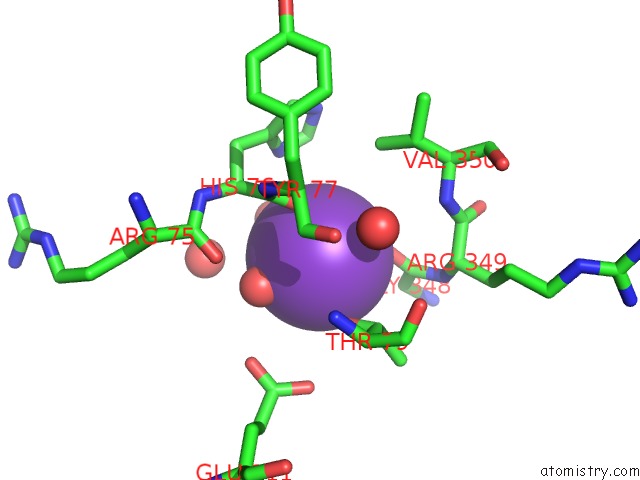
Mono view
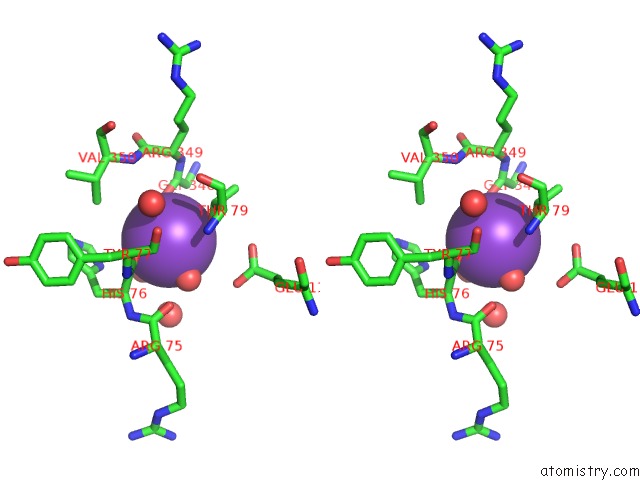
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Crystal Structure of the GRP1 pH Domain in Complex with ARF6-Gtp within 5.0Å range:
|
Reference:
A.W.Malaby,
B.Van Den Berg,
D.G.Lambright.
Structural Basis For Membrane Recruitment and Allosteric Activation of Cytohesin Family Arf Gtpase Exchange Factors. Proc.Natl.Acad.Sci.Usa V. 110 14213 2013.
ISSN: ISSN 0027-8424
PubMed: 23940353
DOI: 10.1073/PNAS.1301883110
Page generated: Sat Aug 9 07:08:44 2025
ISSN: ISSN 0027-8424
PubMed: 23940353
DOI: 10.1073/PNAS.1301883110
Last articles
Mg in 5DQLMg in 5DR2
Mg in 5DQZ
Mg in 5DQK
Mg in 5DOU
Mg in 5DQH
Mg in 5DQG
Mg in 5DPH
Mg in 5DOS
Mg in 5DOK