Potassium »
PDB 3oyt-3q9b »
3ply »
Potassium in PDB 3ply: Structure of Oxidized P96G Mutant of Amicyanin
Protein crystallography data
The structure of Structure of Oxidized P96G Mutant of Amicyanin, PDB code: 3ply
was solved by
N.Sukumar,
V.L.Davidson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.20 |
| Space group | P 32 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 97.279, 97.279, 109.875, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 23.9 / 29.1 |
Other elements in 3ply:
The structure of Structure of Oxidized P96G Mutant of Amicyanin also contains other interesting chemical elements:
| Copper | (Cu) | 4 atoms |
| Sodium | (Na) | 2 atoms |
Potassium Binding Sites:
The binding sites of Potassium atom in the Structure of Oxidized P96G Mutant of Amicyanin
(pdb code 3ply). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total only one binding site of Potassium was determined in the Structure of Oxidized P96G Mutant of Amicyanin, PDB code: 3ply:
In total only one binding site of Potassium was determined in the Structure of Oxidized P96G Mutant of Amicyanin, PDB code: 3ply:
Potassium binding site 1 out of 1 in 3ply
Go back to
Potassium binding site 1 out
of 1 in the Structure of Oxidized P96G Mutant of Amicyanin
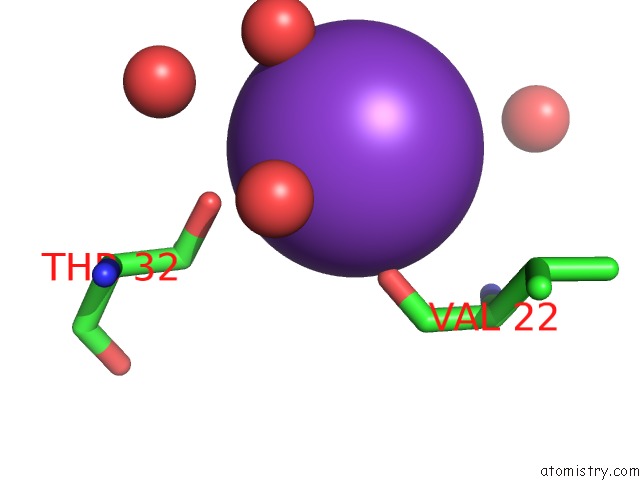
Mono view
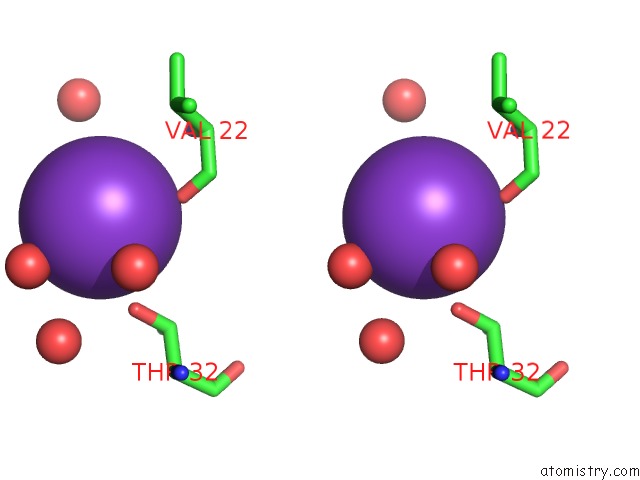
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Structure of Oxidized P96G Mutant of Amicyanin within 5.0Å range:
|
Reference:
M.Choi,
N.Sukumar,
F.S.Mathews,
A.Liu,
V.L.Davidson.
Proline 96 of the Copper Ligand Loop of Amicyanin Regulates Electron Transfer From Methylamine Dehydrogenase By Positioning Other Residues at the Protein-Protein Interface. Biochemistry V. 50 1265 2011.
ISSN: ISSN 0006-2960
PubMed: 21268585
DOI: 10.1021/BI101794Y
Page generated: Mon Aug 12 09:02:39 2024
ISSN: ISSN 0006-2960
PubMed: 21268585
DOI: 10.1021/BI101794Y
Last articles
I in 6W35I in 6W0U
I in 6W42
I in 6W2C
I in 6UIO
I in 6VX2
I in 6VPE
I in 6VU4
I in 6VQS
I in 6VRL