Potassium »
PDB 2o8l-2qxl »
2pur »
Potassium in PDB 2pur: Structure of Dihydrodipicolinate Synthase Mutant THR44SER at 1.7 A.
Enzymatic activity of Structure of Dihydrodipicolinate Synthase Mutant THR44SER at 1.7 A.
All present enzymatic activity of Structure of Dihydrodipicolinate Synthase Mutant THR44SER at 1.7 A.:
4.2.1.52;
4.2.1.52;
Protein crystallography data
The structure of Structure of Dihydrodipicolinate Synthase Mutant THR44SER at 1.7 A., PDB code: 2pur
was solved by
R.C.J.Dobson,
G.B.Jameson,
J.A.Gerrard,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 33.31 / 1.70 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 121.040, 121.040, 110.319, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 17.9 / 20.7 |
Potassium Binding Sites:
The binding sites of Potassium atom in the Structure of Dihydrodipicolinate Synthase Mutant THR44SER at 1.7 A.
(pdb code 2pur). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 2 binding sites of Potassium where determined in the Structure of Dihydrodipicolinate Synthase Mutant THR44SER at 1.7 A., PDB code: 2pur:
Jump to Potassium binding site number: 1; 2;
In total 2 binding sites of Potassium where determined in the Structure of Dihydrodipicolinate Synthase Mutant THR44SER at 1.7 A., PDB code: 2pur:
Jump to Potassium binding site number: 1; 2;
Potassium binding site 1 out of 2 in 2pur
Go back to
Potassium binding site 1 out
of 2 in the Structure of Dihydrodipicolinate Synthase Mutant THR44SER at 1.7 A.
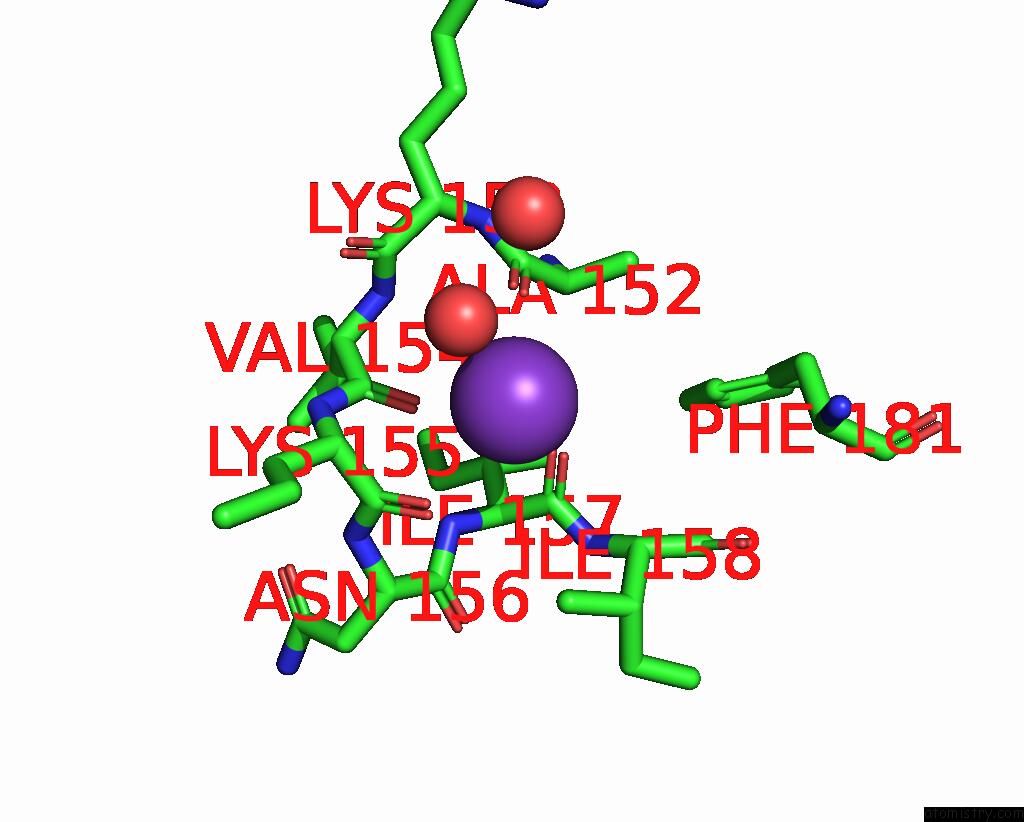
Mono view
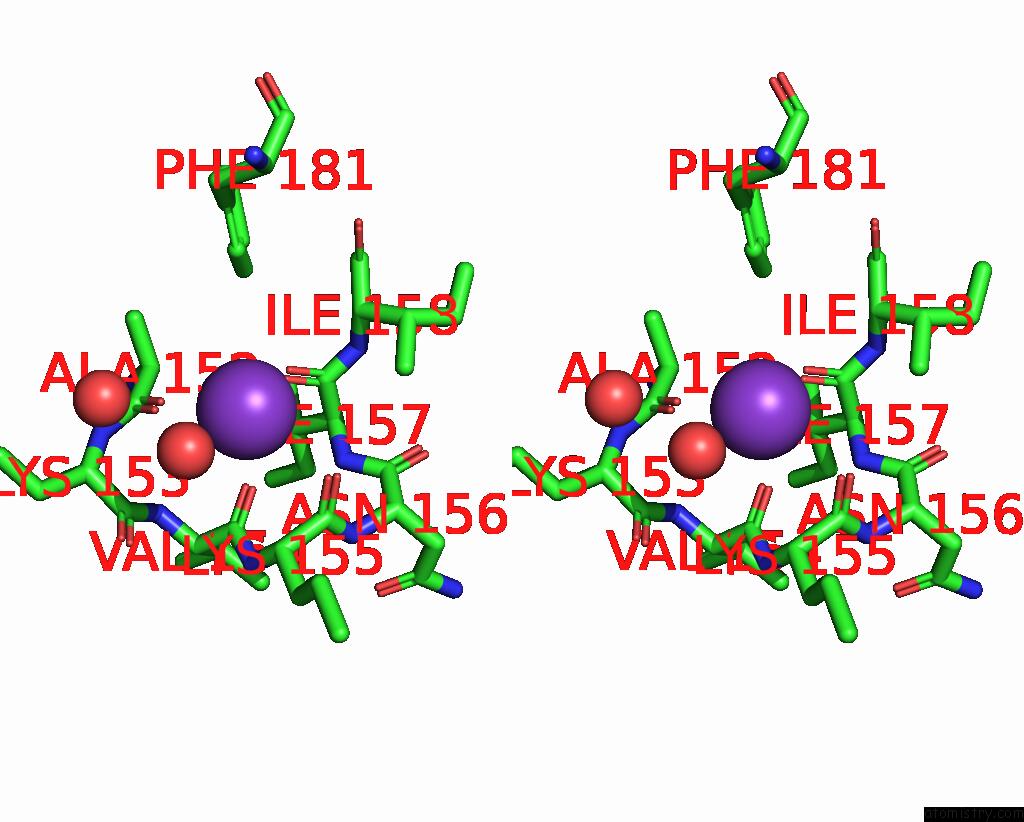
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Structure of Dihydrodipicolinate Synthase Mutant THR44SER at 1.7 A. within 5.0Å range:
|
Potassium binding site 2 out of 2 in 2pur
Go back to
Potassium binding site 2 out
of 2 in the Structure of Dihydrodipicolinate Synthase Mutant THR44SER at 1.7 A.
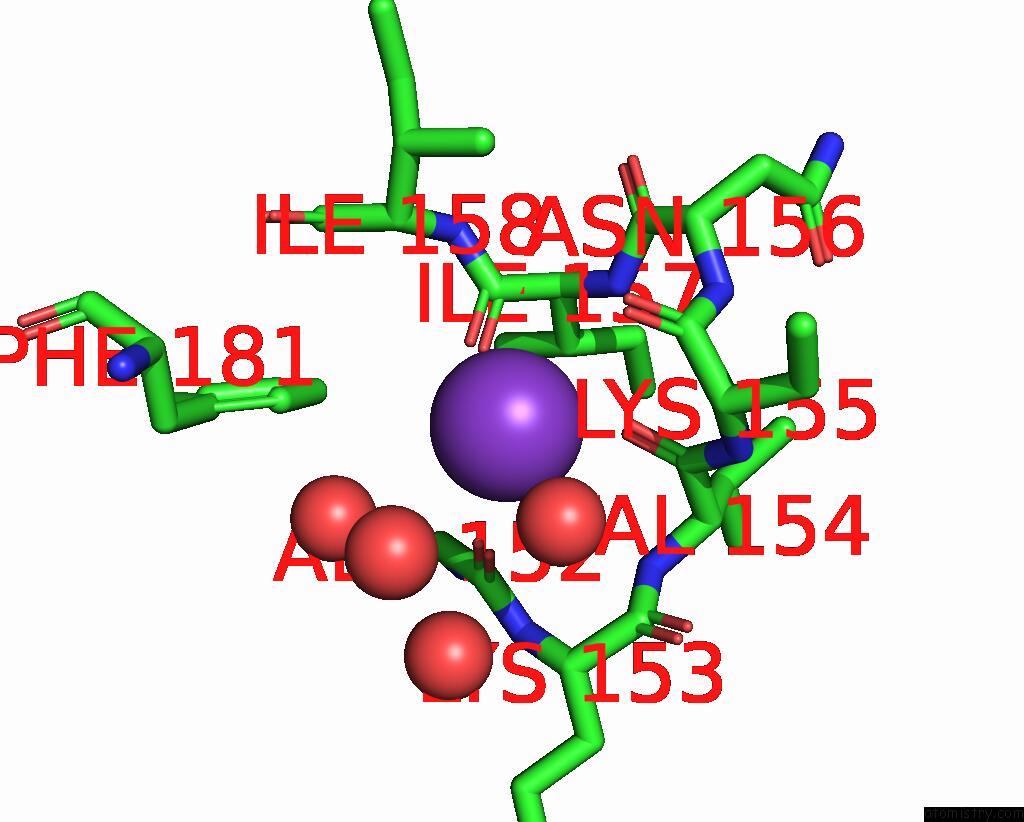
Mono view
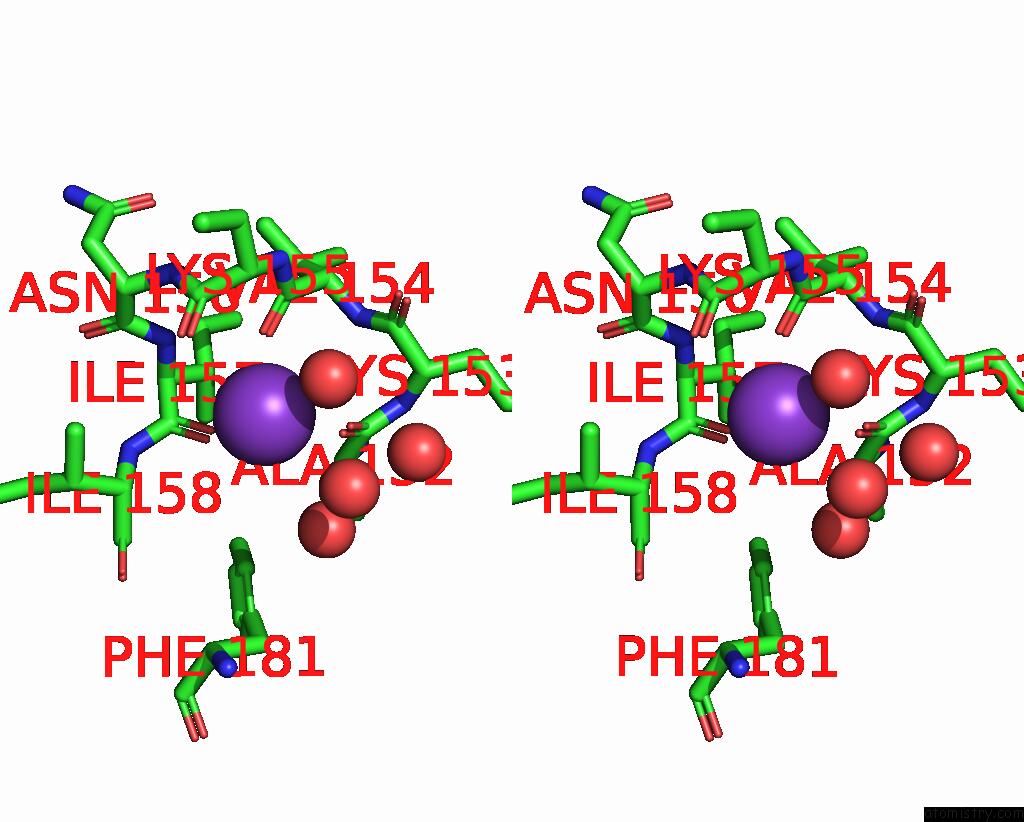
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Structure of Dihydrodipicolinate Synthase Mutant THR44SER at 1.7 A. within 5.0Å range:
|
Reference:
R.C.Dobson,
M.A.Perugini,
G.B.Jameson,
J.A.Gerrard.
Specificity Versus Catalytic Potency: the Role of Threonine 44 in Escherichia Coli Dihydrodipicolinate Synthase Mediated Catalysis. Biochimie V. 91 1036 2009.
ISSN: ISSN 0300-9084
PubMed: 19505526
DOI: 10.1016/J.BIOCHI.2009.05.013
Page generated: Sat Aug 9 03:46:59 2025
ISSN: ISSN 0300-9084
PubMed: 19505526
DOI: 10.1016/J.BIOCHI.2009.05.013
Last articles
Na in 4CNPNa in 4CNN
Na in 4CKY
Na in 4CNT
Na in 4CDB
Na in 4CNC
Na in 4CK4
Na in 4CIW
Na in 4CIY
Na in 4CIT