Potassium »
PDB 9gku-9mek »
9j0x »
Potassium in PDB 9j0x: Cryo-Em Structure of the Guard Cell Potassium Channel Gork
Potassium Binding Sites:
The binding sites of Potassium atom in the Cryo-Em Structure of the Guard Cell Potassium Channel Gork
(pdb code 9j0x). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 4 binding sites of Potassium where determined in the Cryo-Em Structure of the Guard Cell Potassium Channel Gork, PDB code: 9j0x:
Jump to Potassium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Potassium where determined in the Cryo-Em Structure of the Guard Cell Potassium Channel Gork, PDB code: 9j0x:
Jump to Potassium binding site number: 1; 2; 3; 4;
Potassium binding site 1 out of 4 in 9j0x
Go back to
Potassium binding site 1 out
of 4 in the Cryo-Em Structure of the Guard Cell Potassium Channel Gork
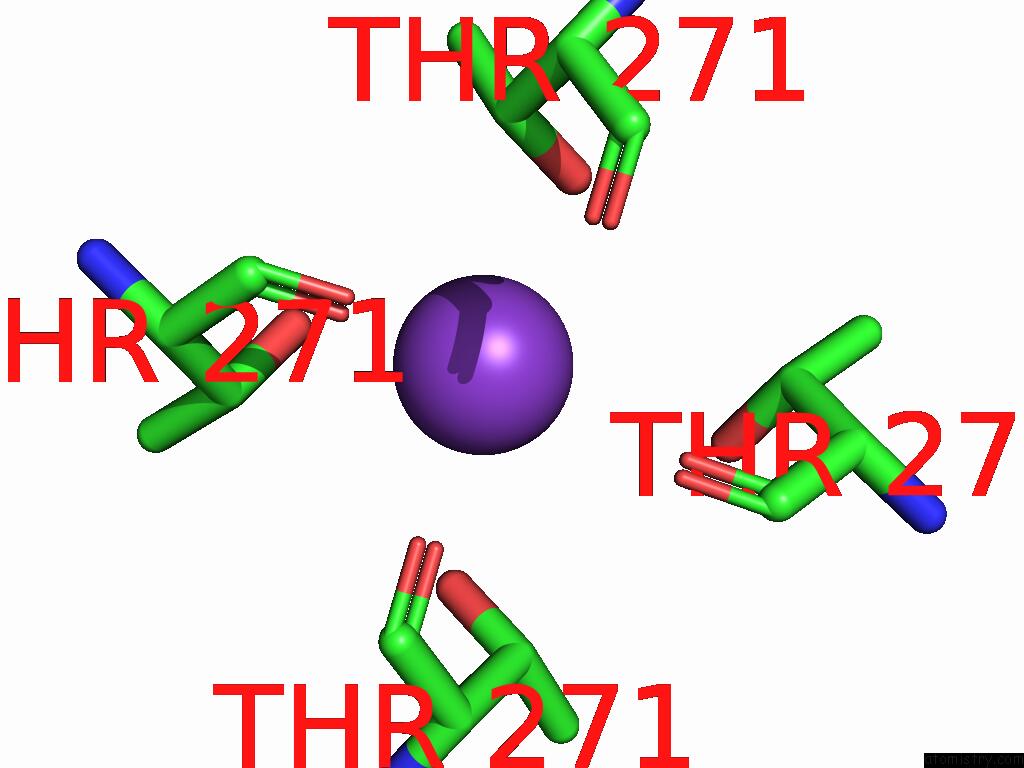
Mono view
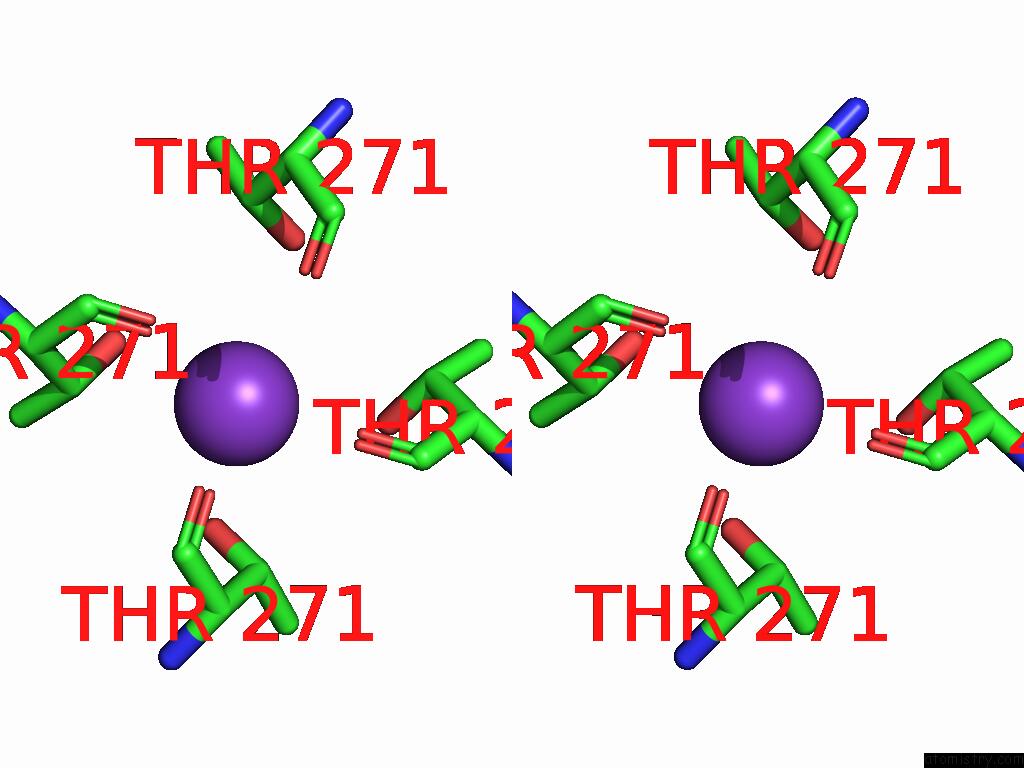
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Cryo-Em Structure of the Guard Cell Potassium Channel Gork within 5.0Å range:
|
Potassium binding site 2 out of 4 in 9j0x
Go back to
Potassium binding site 2 out
of 4 in the Cryo-Em Structure of the Guard Cell Potassium Channel Gork
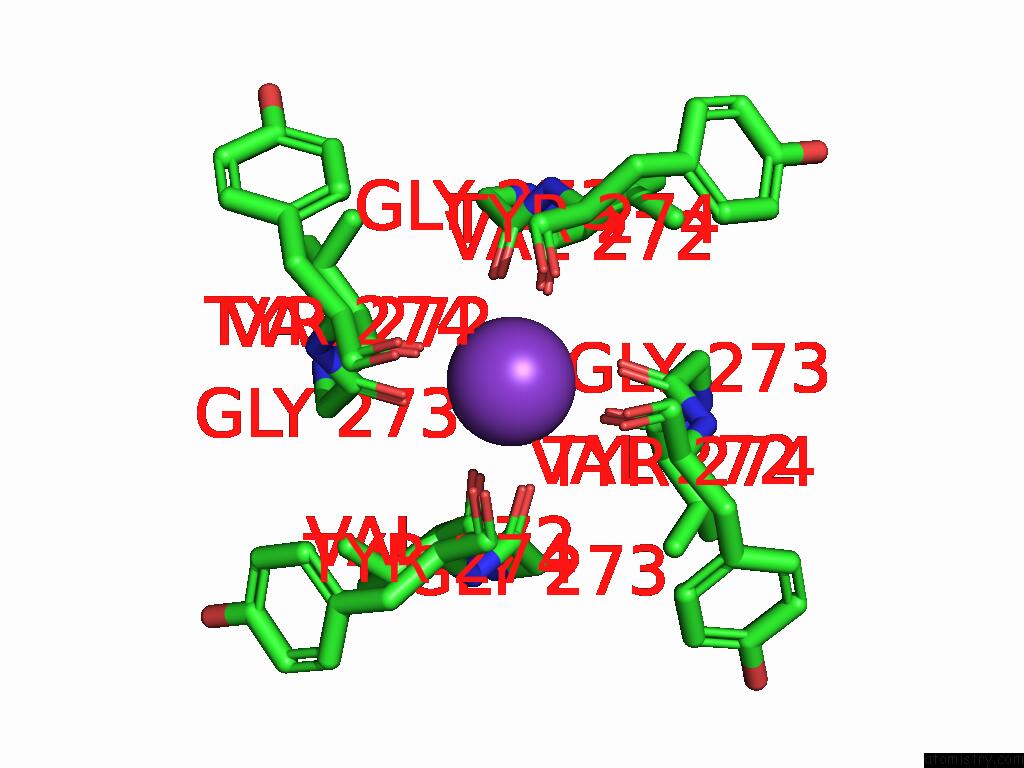
Mono view
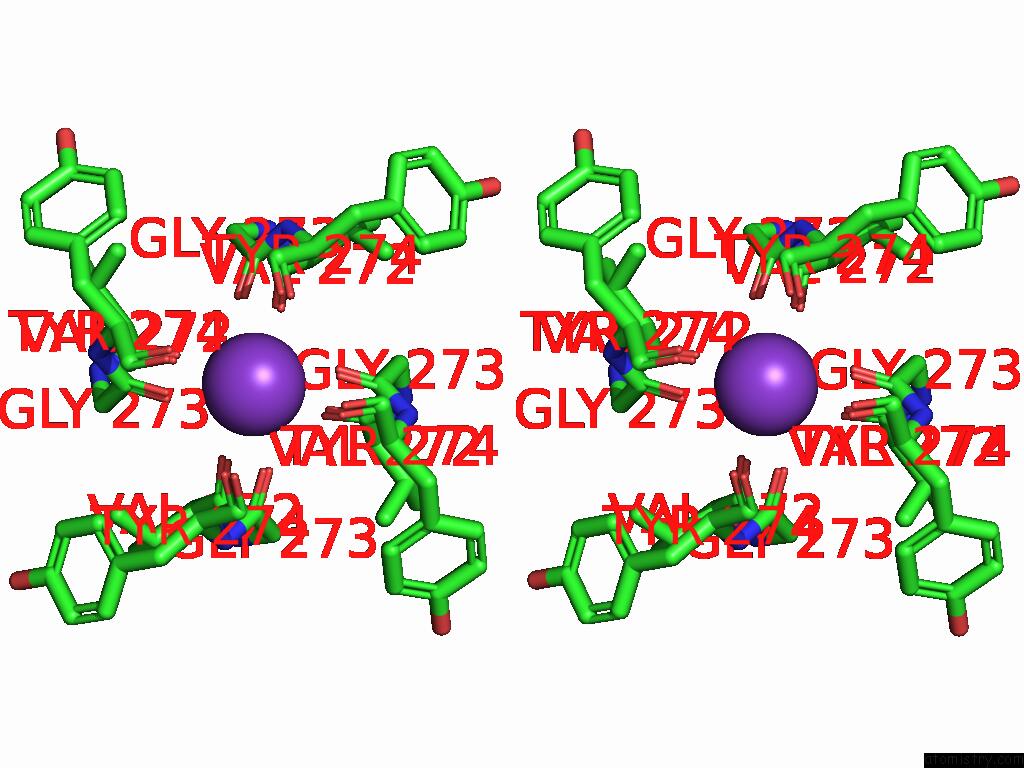
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Cryo-Em Structure of the Guard Cell Potassium Channel Gork within 5.0Å range:
|
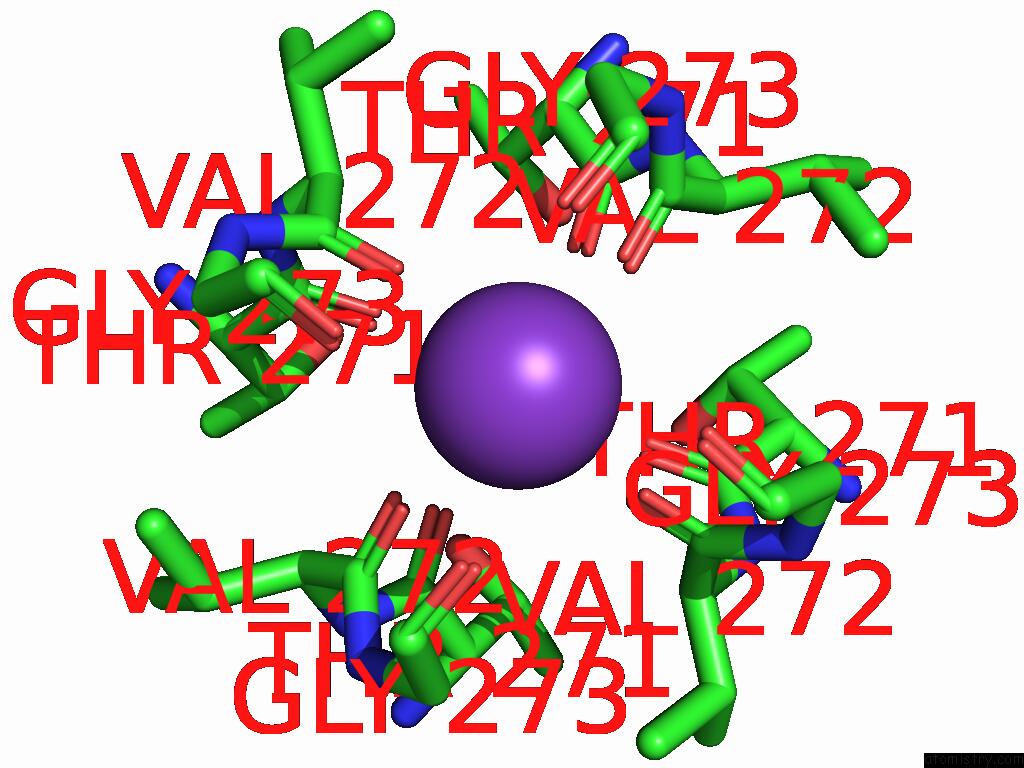
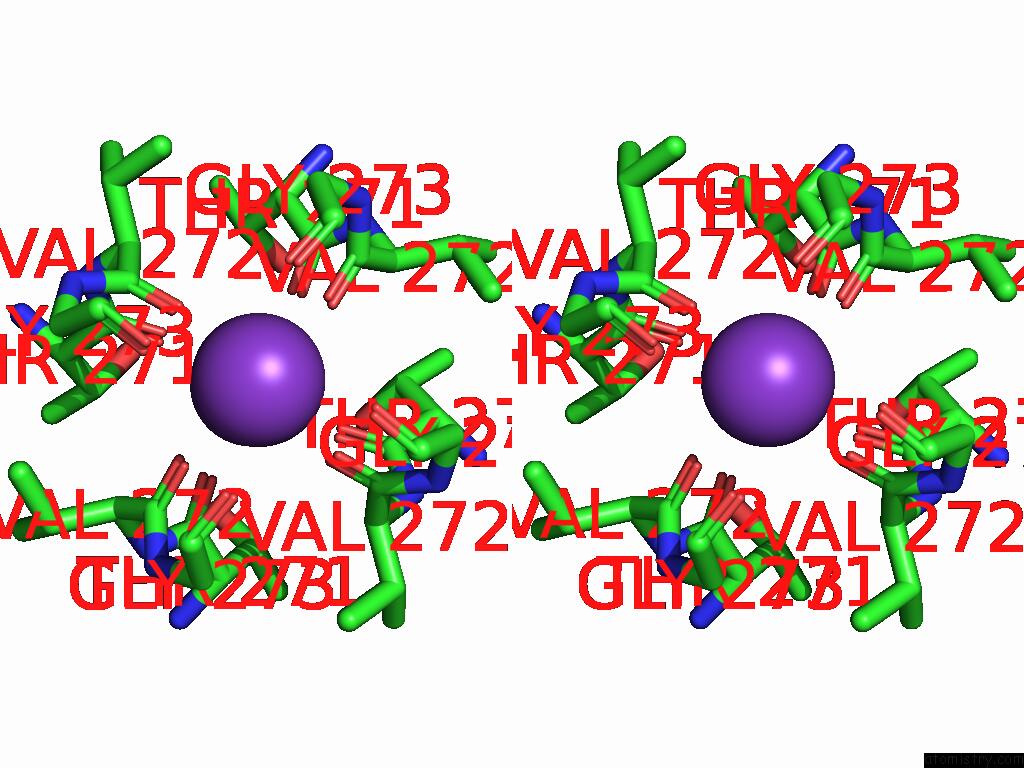
Potassium binding site 3 out of 4 in 9j0x
Go back to
Potassium binding site 3 out
of 4 in the Cryo-Em Structure of the Guard Cell Potassium Channel Gork
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 3 of Cryo-Em Structure of the Guard Cell Potassium Channel Gork within 5.0Å range:
|
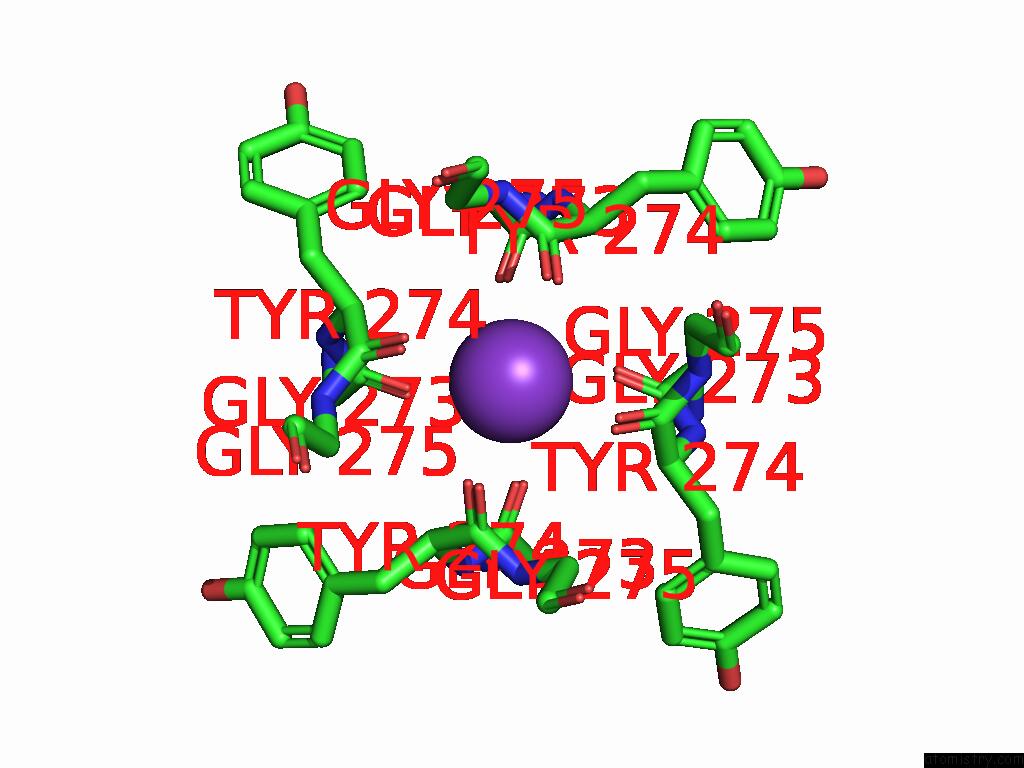
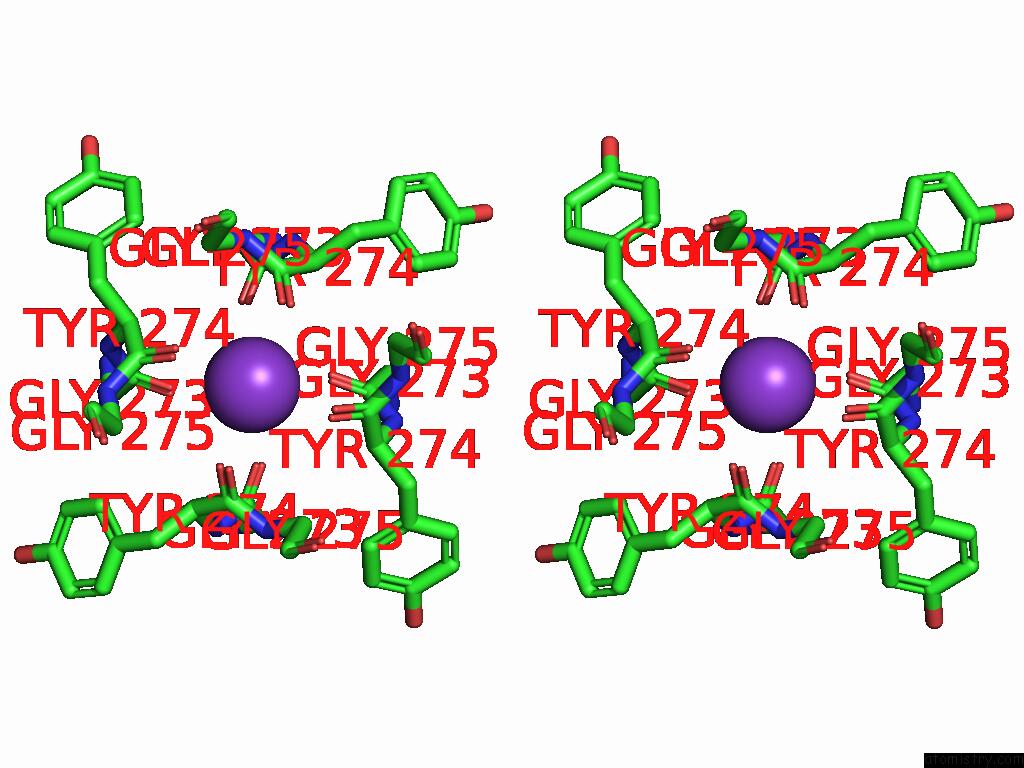
Potassium binding site 4 out of 4 in 9j0x
Go back to
Potassium binding site 4 out
of 4 in the Cryo-Em Structure of the Guard Cell Potassium Channel Gork
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 4 of Cryo-Em Structure of the Guard Cell Potassium Channel Gork within 5.0Å range:
|
Reference:
X.Zhang,
P.Zhang.
Gork K+ Channel Structure and Gating Vital For Stomatal Movements To Be Published.
Page generated: Sat Aug 9 19:14:47 2025
Last articles
Mg in 5JWAMg in 5JVL
Mg in 5JVN
Mg in 5JVM
Mg in 5JVJ
Mg in 5JVD
Mg in 5JV5
Mg in 5JTG
Mg in 5JRW
Mg in 5JSQ