Potassium »
PDB 9gku-9mek »
9i0m »
Potassium in PDB 9i0m: Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended
Enzymatic activity of Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended
All present enzymatic activity of Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended:
1.1.1.205;
1.1.1.205;
Potassium Binding Sites:
The binding sites of Potassium atom in the Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended
(pdb code 9i0m). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 8 binding sites of Potassium where determined in the Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended, PDB code: 9i0m:
Jump to Potassium binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Potassium where determined in the Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended, PDB code: 9i0m:
Jump to Potassium binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Potassium binding site 1 out of 8 in 9i0m
Go back to
Potassium binding site 1 out
of 8 in the Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended
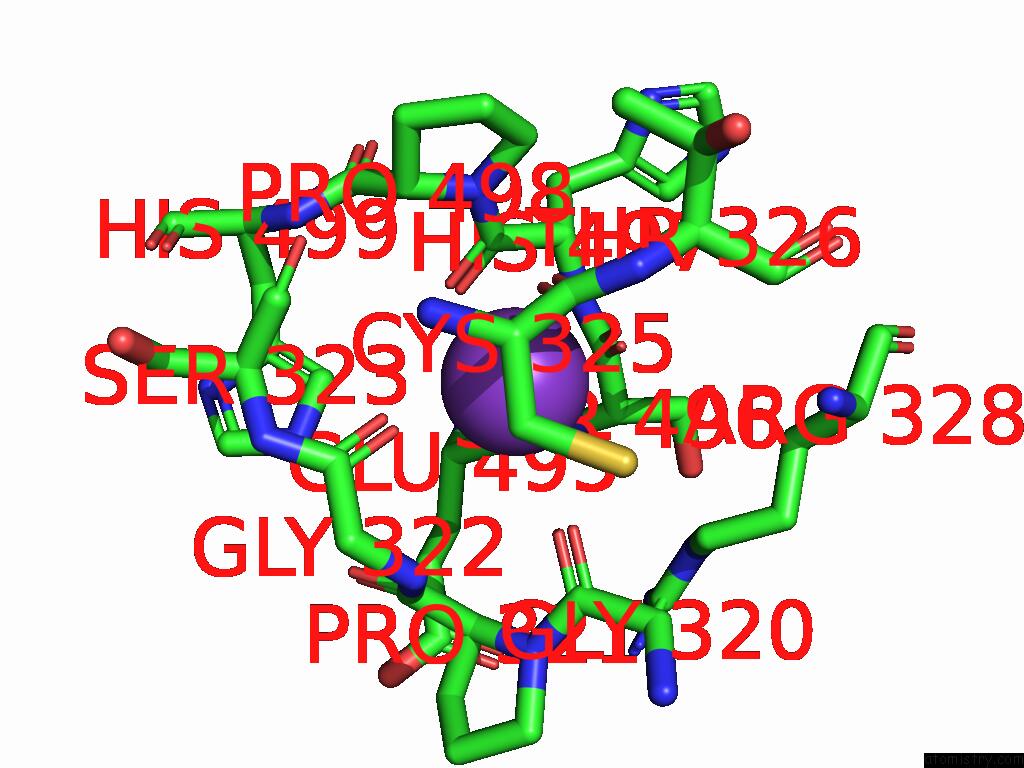
Mono view
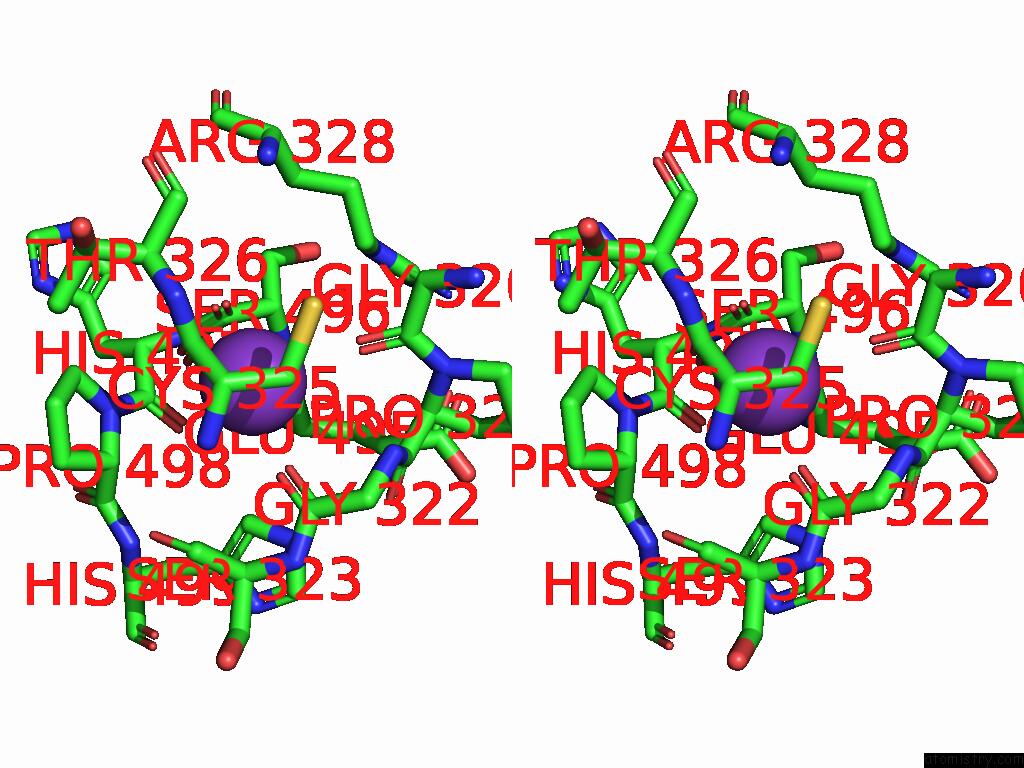
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended within 5.0Å range:
|
Potassium binding site 2 out of 8 in 9i0m
Go back to
Potassium binding site 2 out
of 8 in the Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended
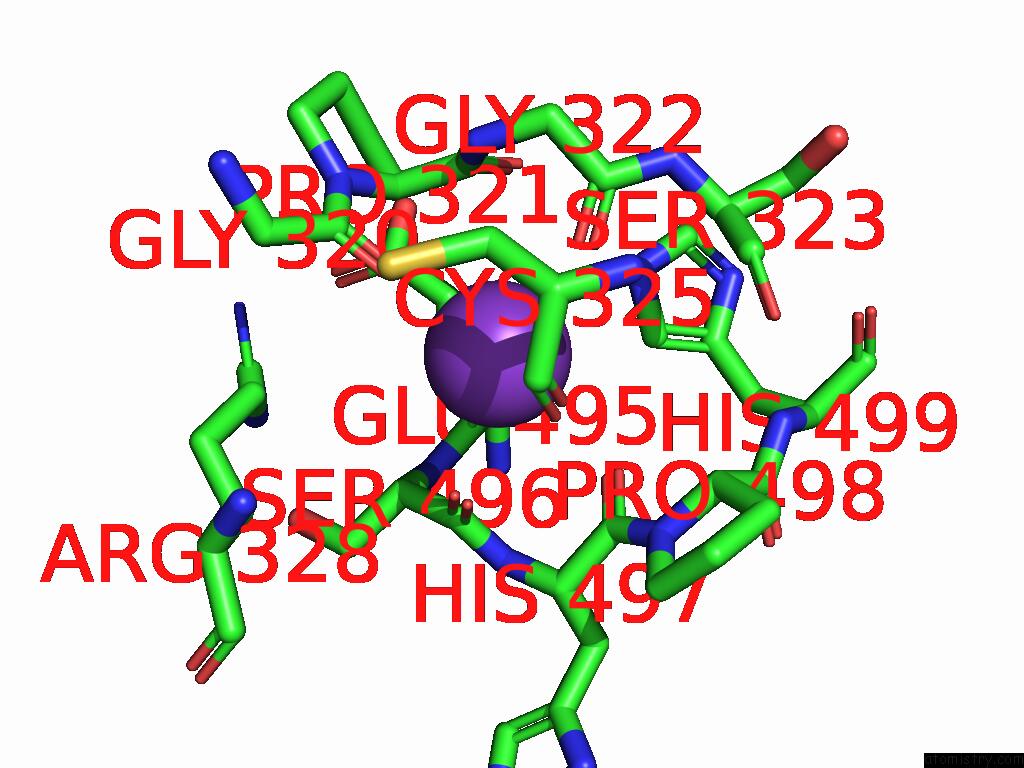
Mono view
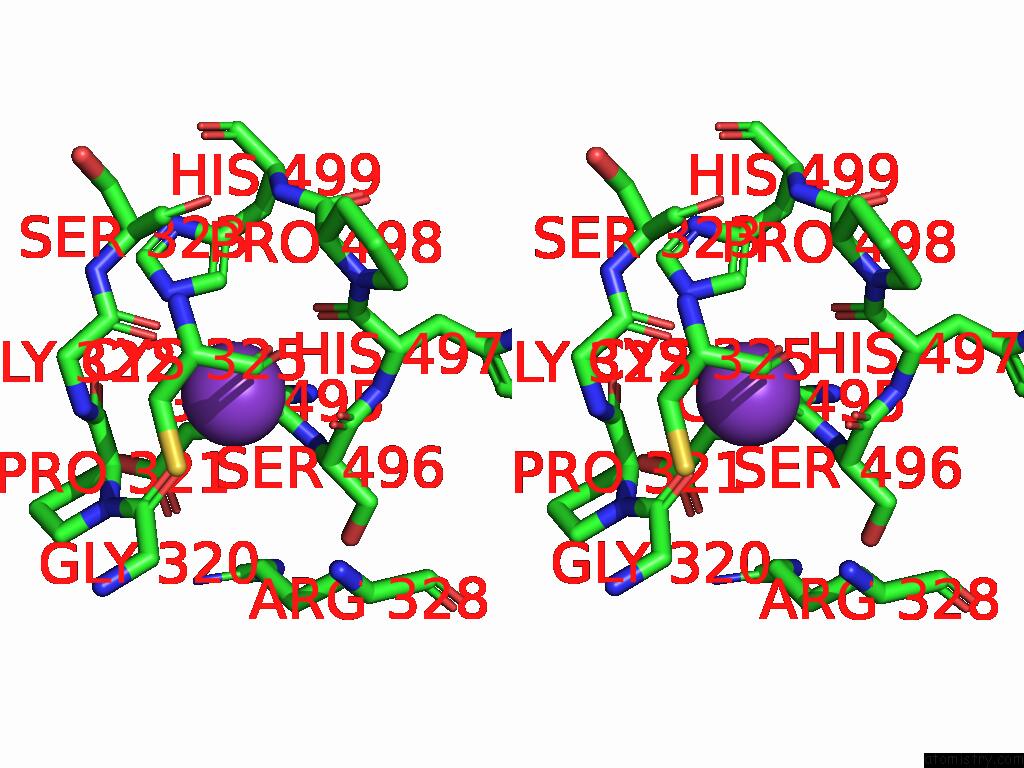
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended within 5.0Å range:
|
Potassium binding site 3 out of 8 in 9i0m
Go back to
Potassium binding site 3 out
of 8 in the Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended
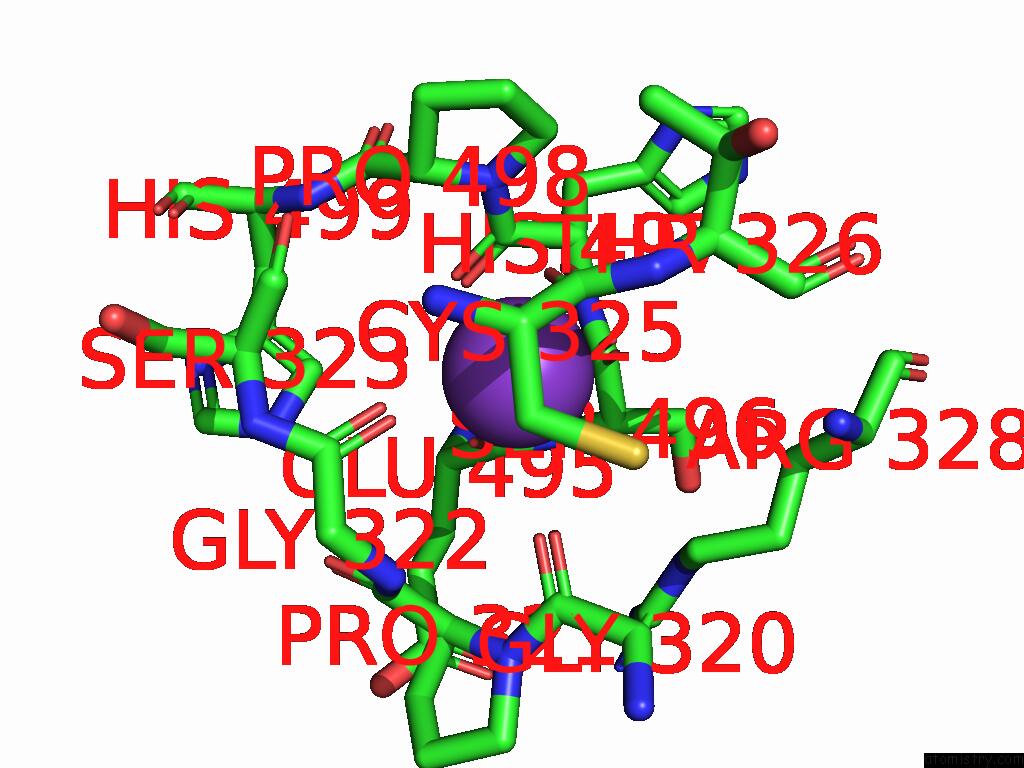
Mono view
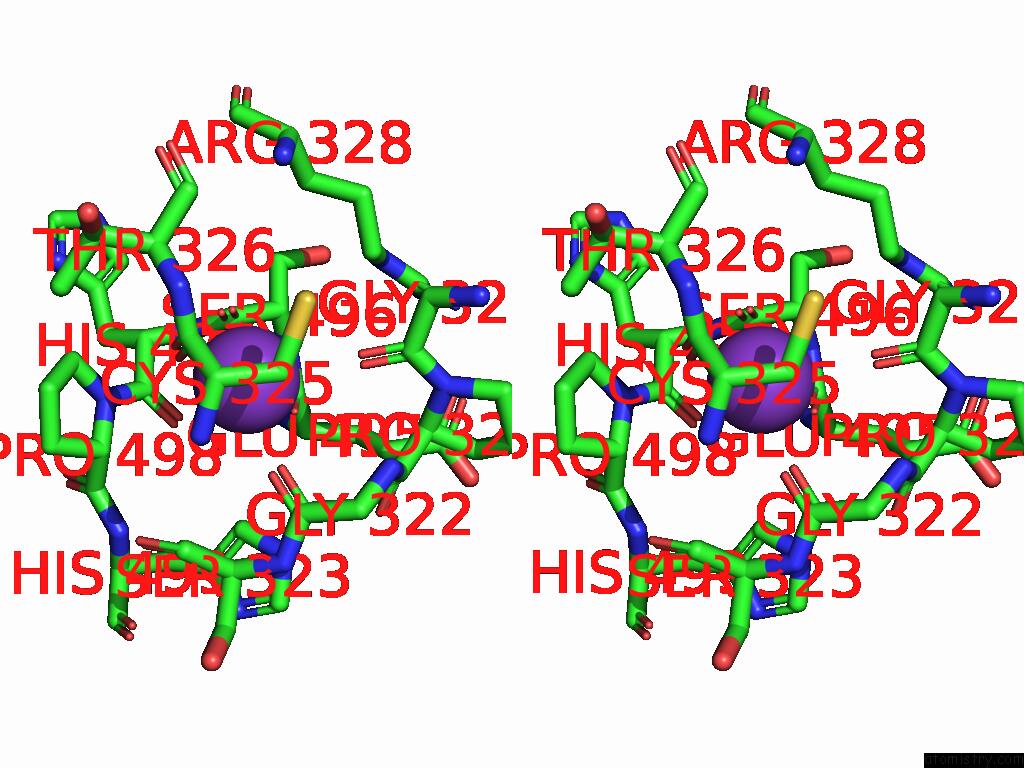
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 3 of Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended within 5.0Å range:
|
Potassium binding site 4 out of 8 in 9i0m
Go back to
Potassium binding site 4 out
of 8 in the Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended
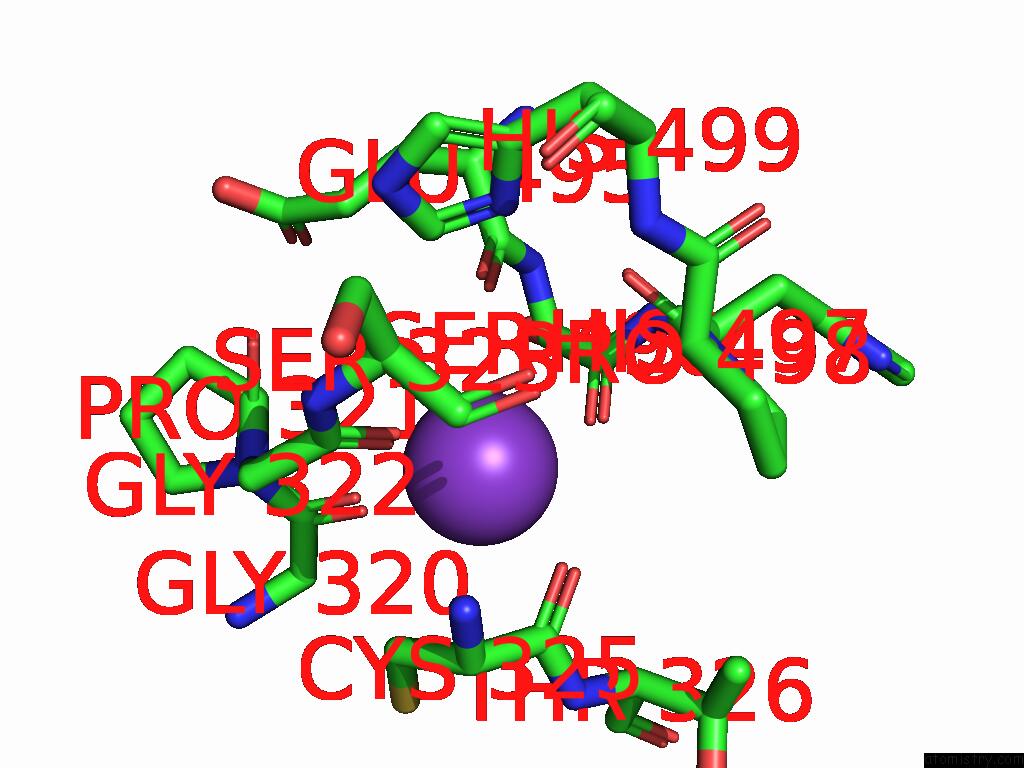
Mono view
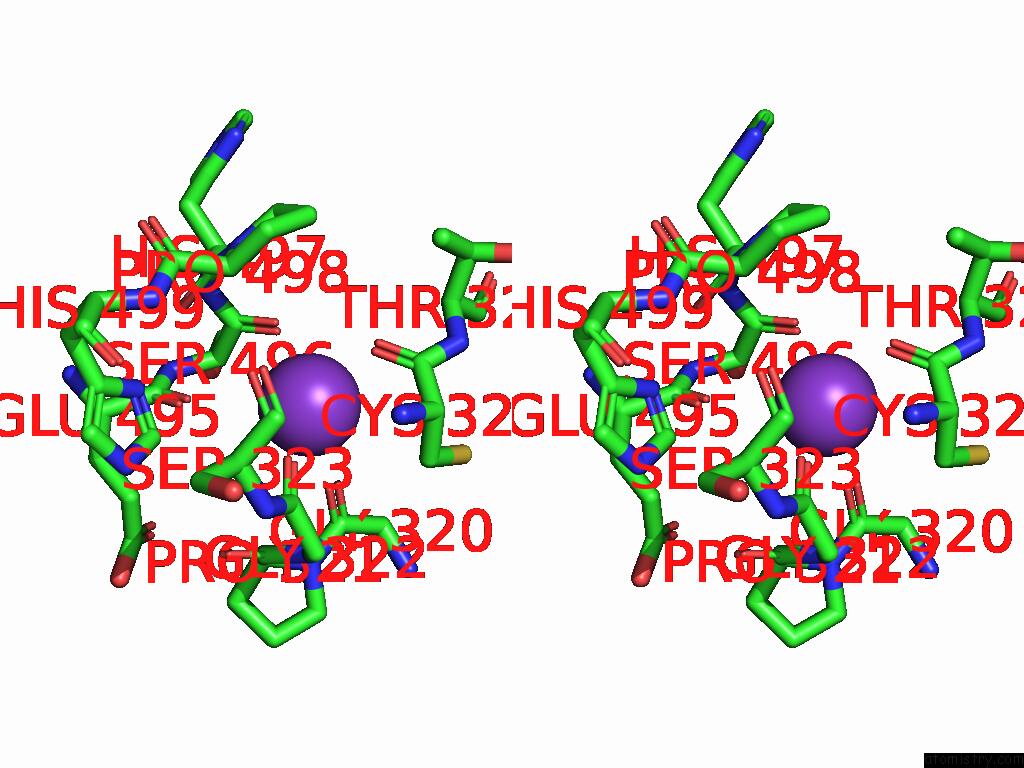
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 4 of Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended within 5.0Å range:
|
Potassium binding site 5 out of 8 in 9i0m
Go back to
Potassium binding site 5 out
of 8 in the Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended
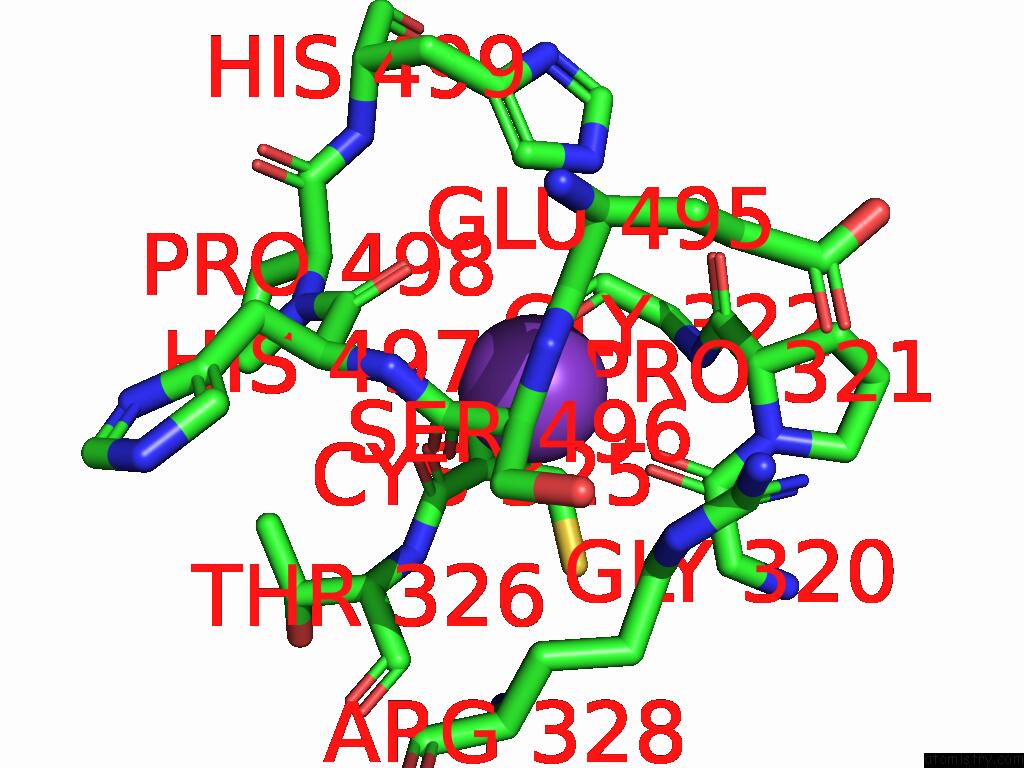
Mono view
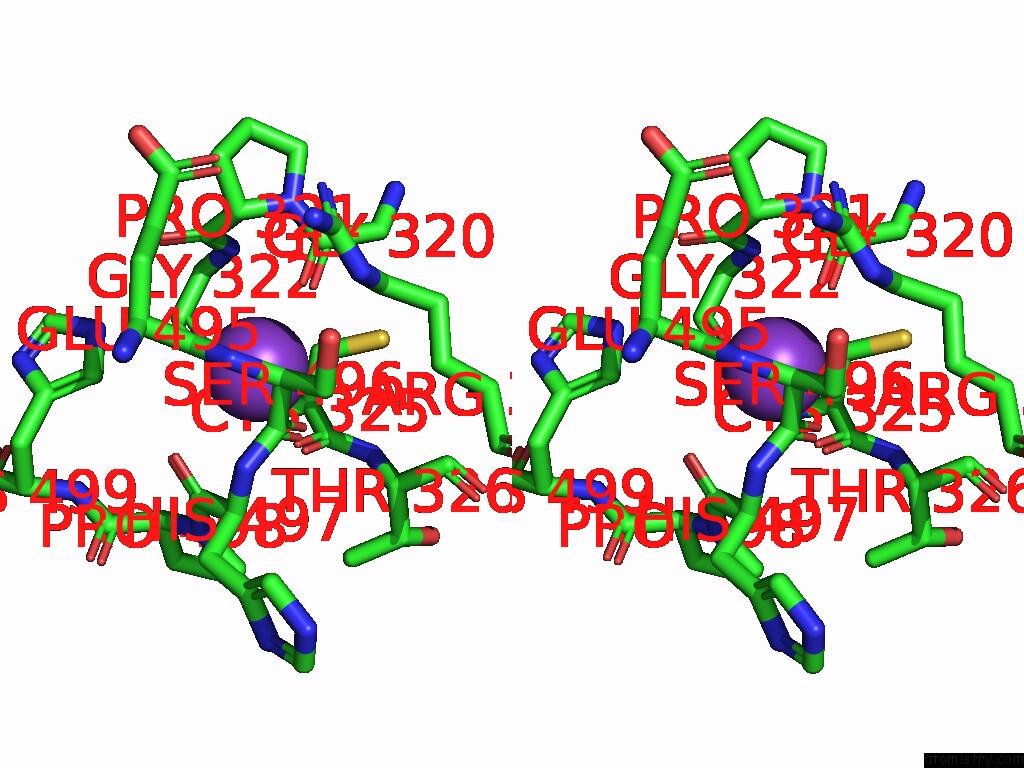
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 5 of Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended within 5.0Å range:
|
Potassium binding site 6 out of 8 in 9i0m
Go back to
Potassium binding site 6 out
of 8 in the Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended
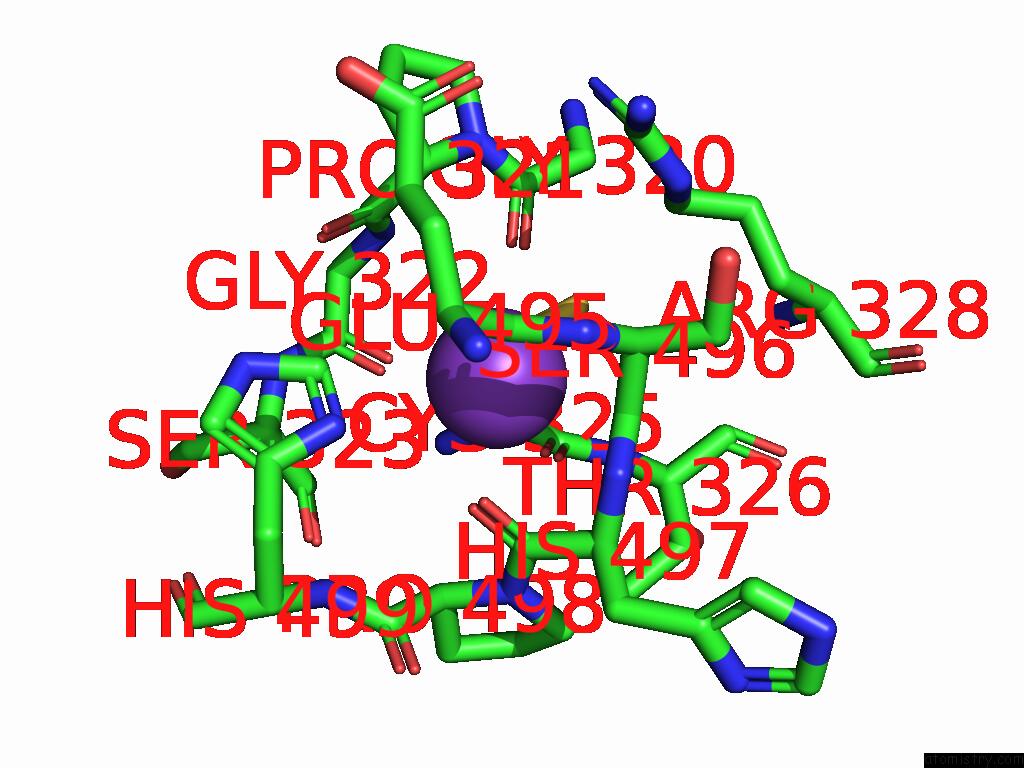
Mono view
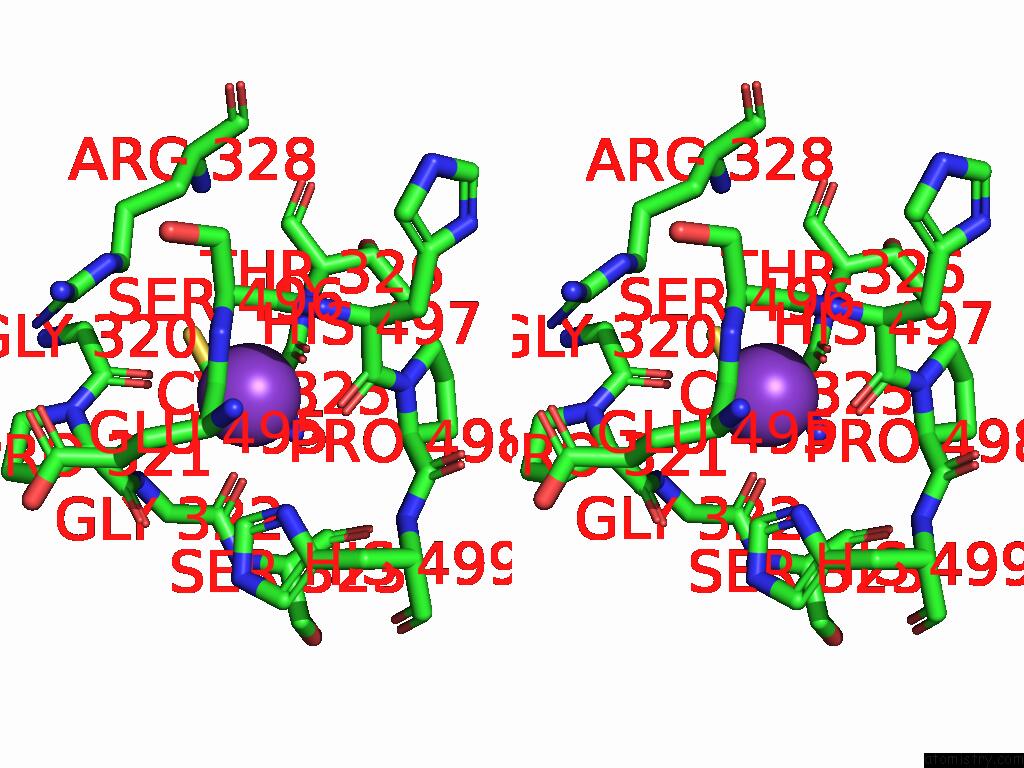
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 6 of Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended within 5.0Å range:
|
Potassium binding site 7 out of 8 in 9i0m
Go back to
Potassium binding site 7 out
of 8 in the Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended
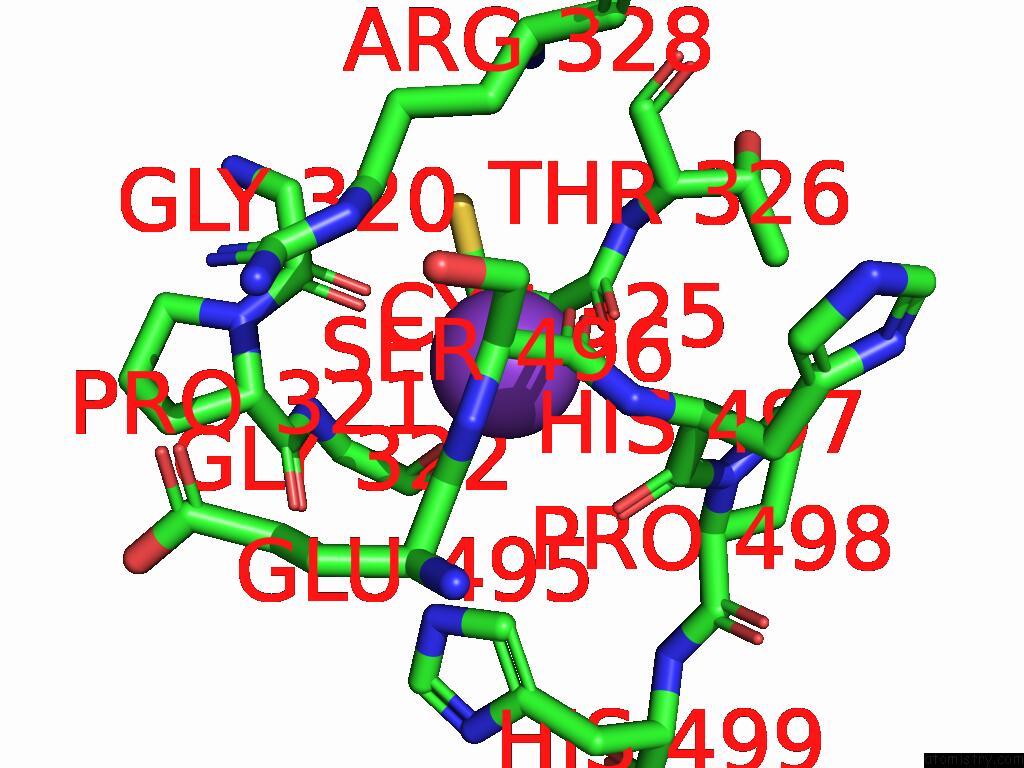
Mono view
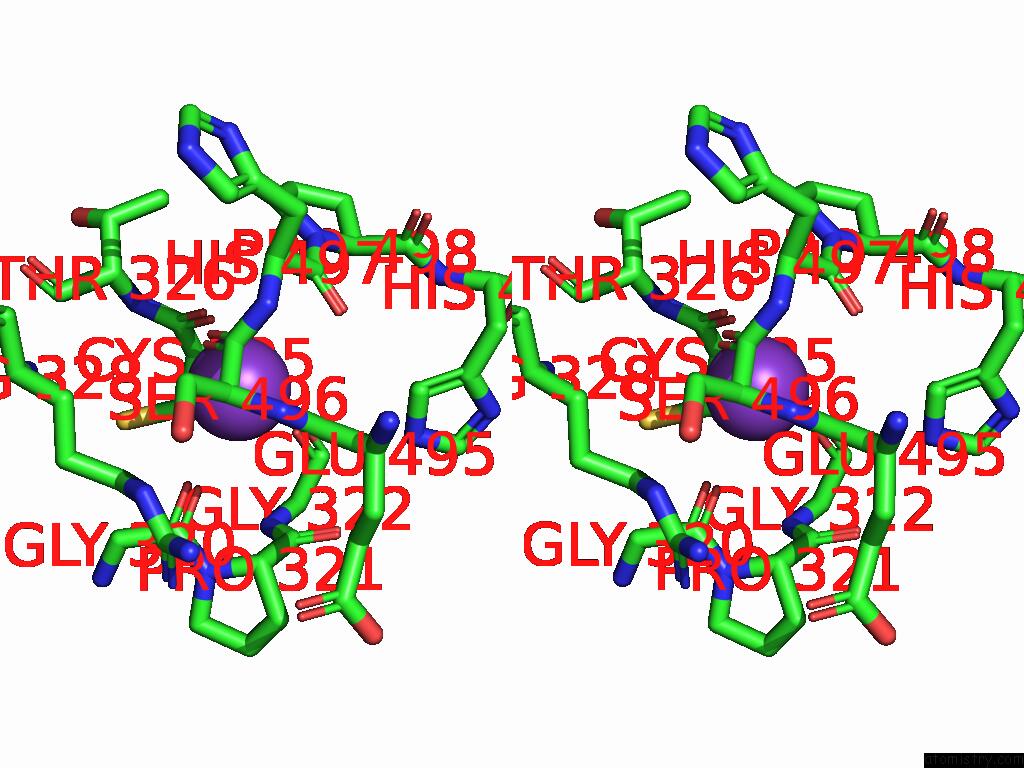
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 7 of Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended within 5.0Å range:
|
Potassium binding site 8 out of 8 in 9i0m
Go back to
Potassium binding site 8 out
of 8 in the Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended
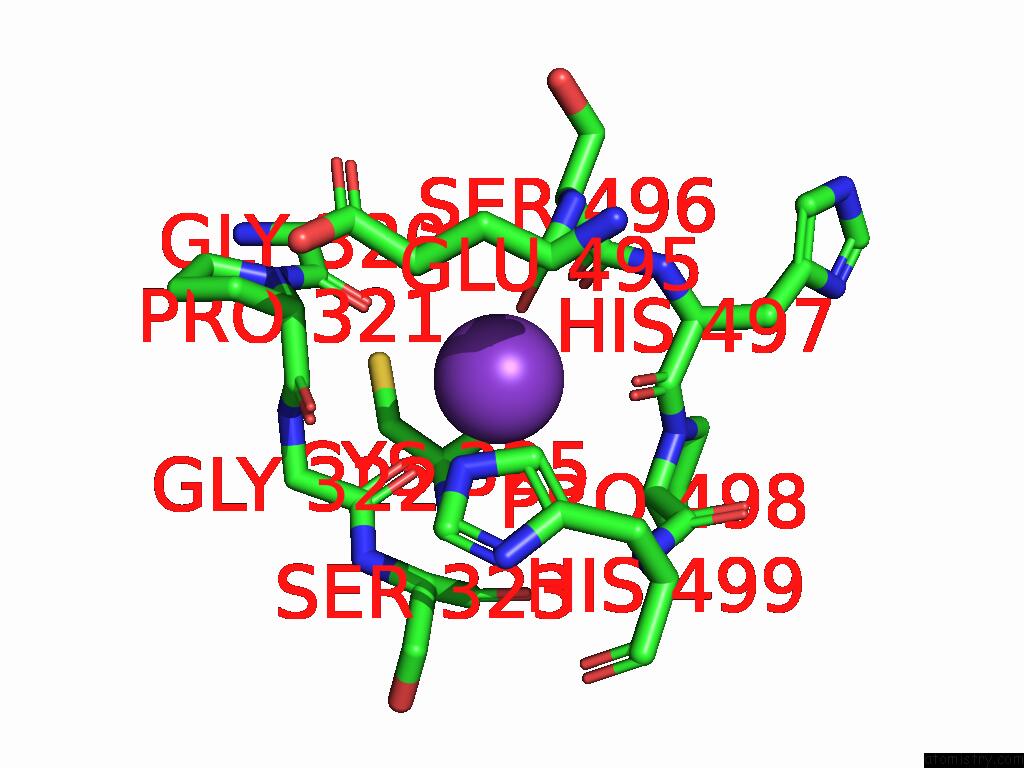
Mono view
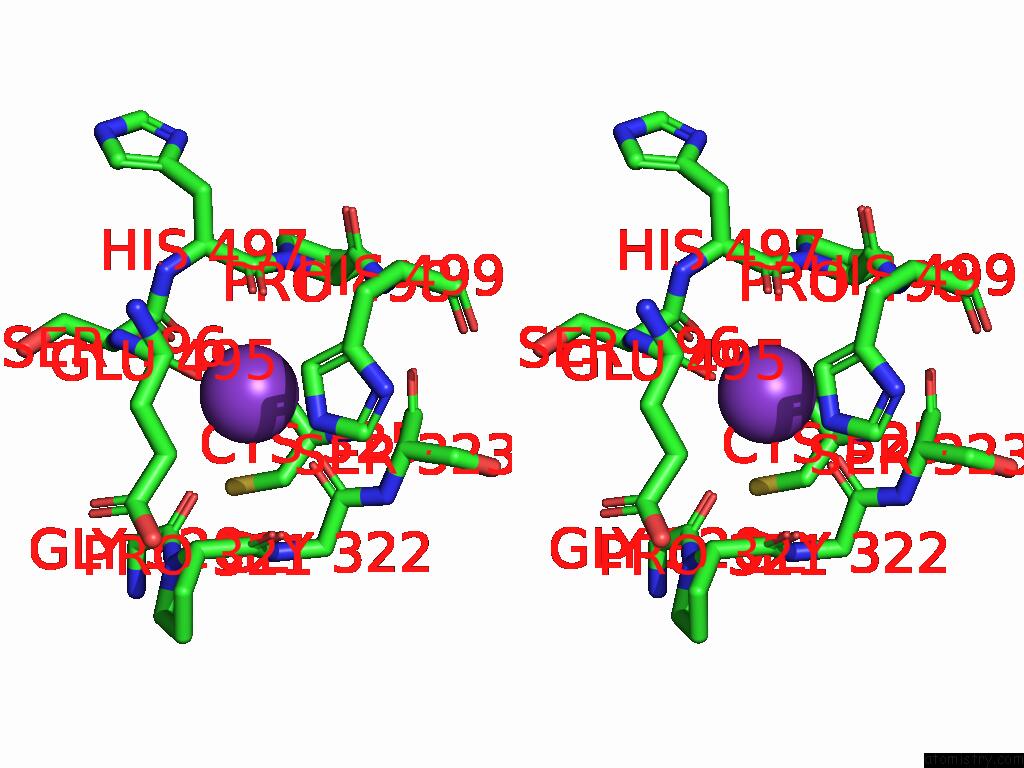
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 8 of Mycobacterium Smegmatis Inosine Monophosphate Dehydrogenase (Impdh) Saturating Atp+Imp-Bound Form, Extended within 5.0Å range:
|
Reference:
O.Bulvas,
Z.Knejzlik,
A.Filimonenko,
T.Kouba,
I.Pichova.
Conformational Landscape of the Mycobacterial Inosine 5'-Monophosphate Dehydrogenase Octamerization Interface. J.Struct.Biol. V. 217 08198 2025.
ISSN: ESSN 1095-8657
PubMed: 40107326
DOI: 10.1016/J.JSB.2025.108198
Page generated: Sat Aug 9 19:12:05 2025
ISSN: ESSN 1095-8657
PubMed: 40107326
DOI: 10.1016/J.JSB.2025.108198
Last articles
Mg in 3DYGMg in 3DYN
Mg in 3DYH
Mg in 3DYF
Mg in 3DYL
Mg in 3DVA
Mg in 3DY8
Mg in 3DVL
Mg in 3DY7
Mg in 3DXJ