Potassium »
PDB 8qrk-8sfz »
8sda »
Potassium in PDB 8sda: Cryoem Structure of Rat KV2.1(1-598) L403A Mutant in Nanodiscs
Potassium Binding Sites:
The binding sites of Potassium atom in the Cryoem Structure of Rat KV2.1(1-598) L403A Mutant in Nanodiscs
(pdb code 8sda). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 4 binding sites of Potassium where determined in the Cryoem Structure of Rat KV2.1(1-598) L403A Mutant in Nanodiscs, PDB code: 8sda:
Jump to Potassium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Potassium where determined in the Cryoem Structure of Rat KV2.1(1-598) L403A Mutant in Nanodiscs, PDB code: 8sda:
Jump to Potassium binding site number: 1; 2; 3; 4;
Potassium binding site 1 out of 4 in 8sda
Go back to
Potassium binding site 1 out
of 4 in the Cryoem Structure of Rat KV2.1(1-598) L403A Mutant in Nanodiscs
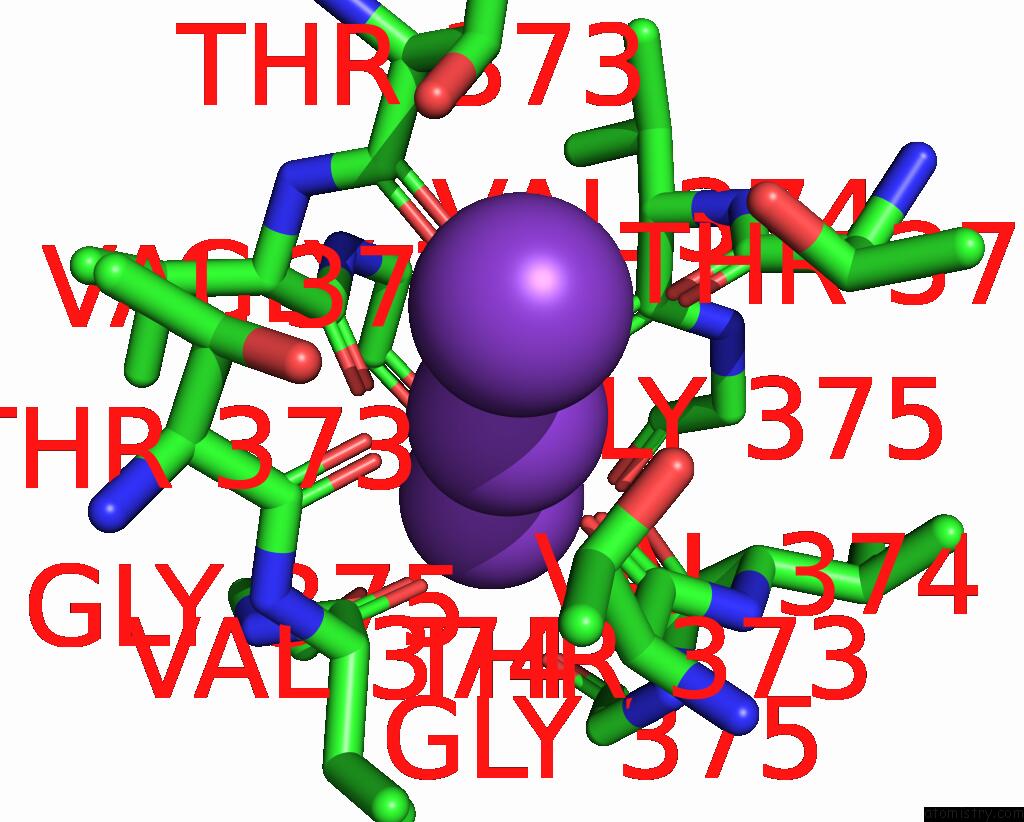
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Cryoem Structure of Rat KV2.1(1-598) L403A Mutant in Nanodiscs within 5.0Å range:
|
Potassium binding site 2 out of 4 in 8sda
Go back to
Potassium binding site 2 out
of 4 in the Cryoem Structure of Rat KV2.1(1-598) L403A Mutant in Nanodiscs
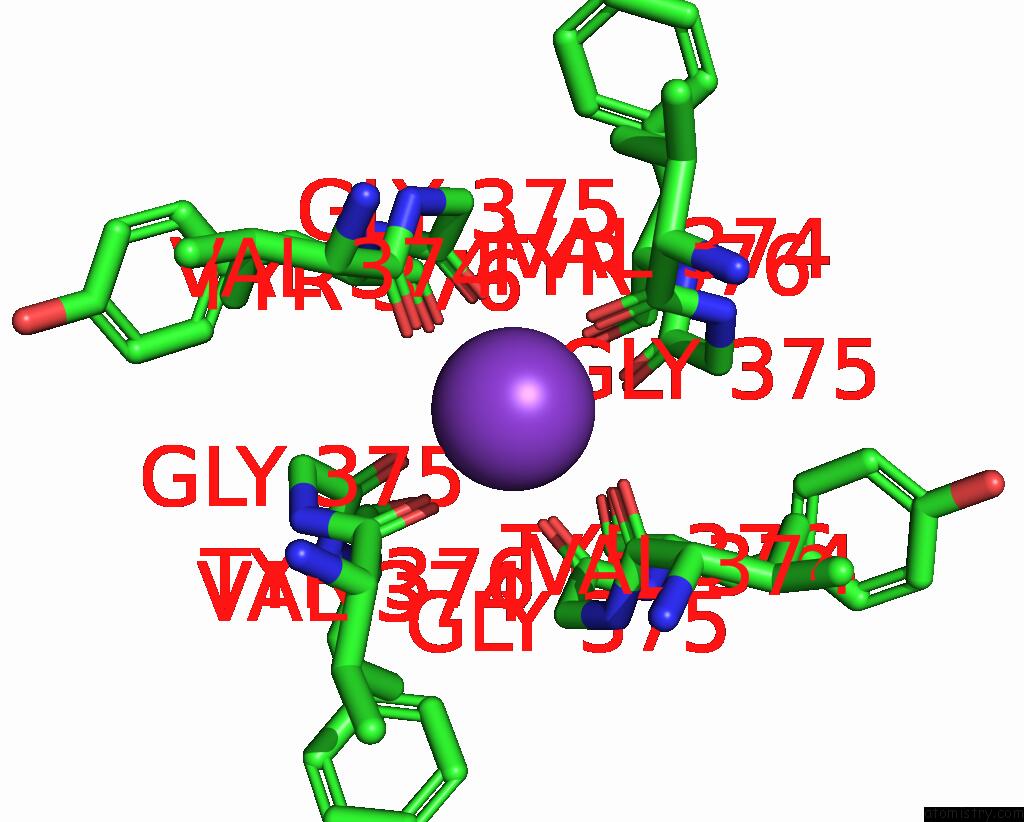
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Cryoem Structure of Rat KV2.1(1-598) L403A Mutant in Nanodiscs within 5.0Å range:
|
Potassium binding site 3 out of 4 in 8sda
Go back to
Potassium binding site 3 out
of 4 in the Cryoem Structure of Rat KV2.1(1-598) L403A Mutant in Nanodiscs
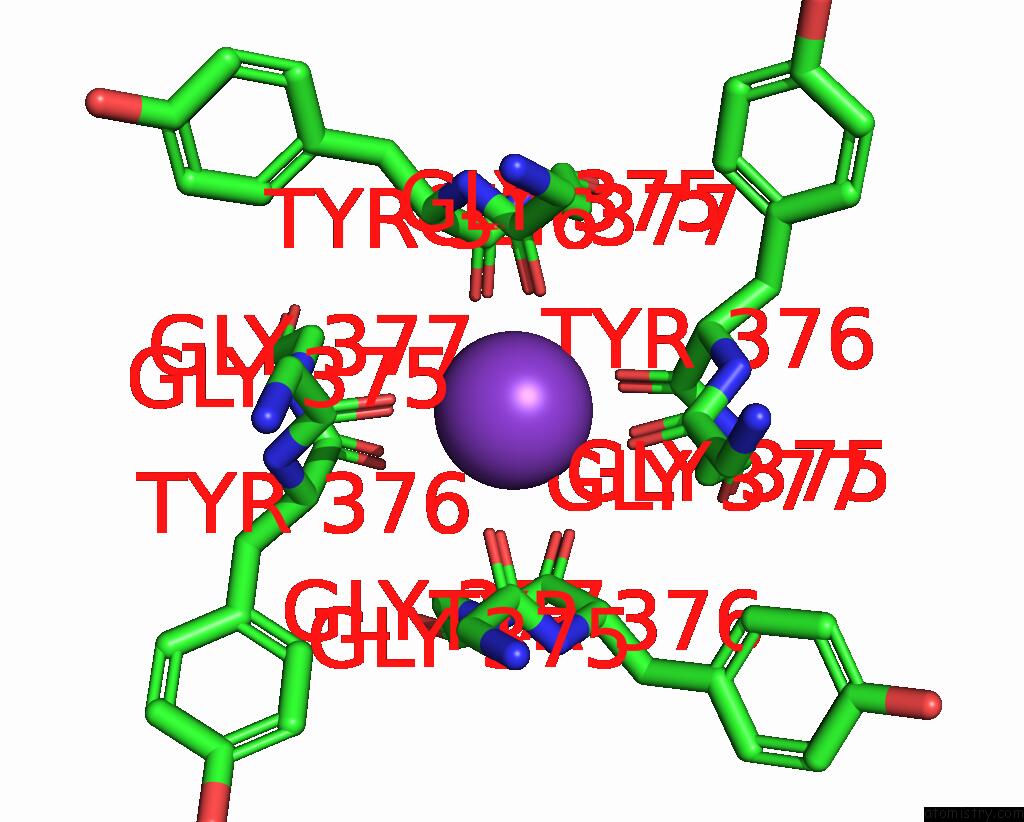
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 3 of Cryoem Structure of Rat KV2.1(1-598) L403A Mutant in Nanodiscs within 5.0Å range:
|
Potassium binding site 4 out of 4 in 8sda
Go back to
Potassium binding site 4 out
of 4 in the Cryoem Structure of Rat KV2.1(1-598) L403A Mutant in Nanodiscs

Mono view
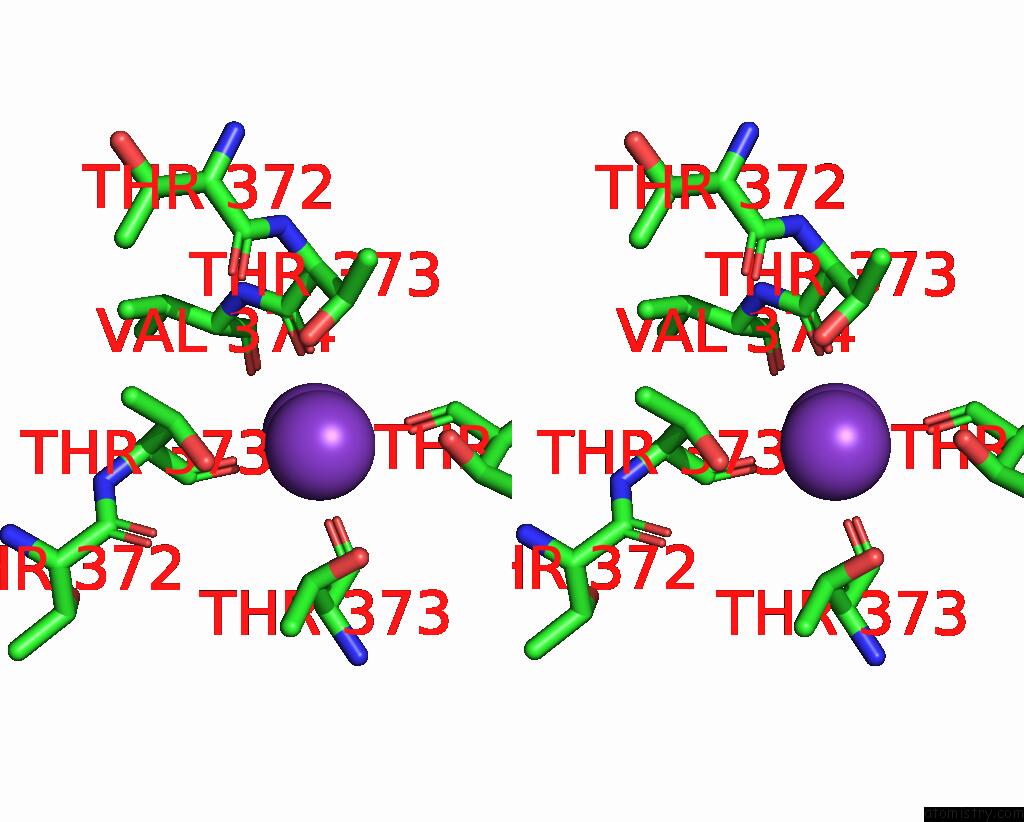
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 4 of Cryoem Structure of Rat KV2.1(1-598) L403A Mutant in Nanodiscs within 5.0Å range:
|
Reference:
A.I.Fernandez-Marino,
X.-F.Tan,
C.Bae,
K.Huffer,
J.Jiang,
K.J.Swartz.
Cryoem Structure of Rat KV2.1(1-598) in Nanodiscs Nature 2023.
ISSN: ESSN 1476-4687
DOI: 10.1038/S41586-023-06582-8
Page generated: Sat Aug 9 17:47:49 2025
ISSN: ESSN 1476-4687
DOI: 10.1038/S41586-023-06582-8
Last articles
Mg in 2X9FMg in 2X9H
Mg in 2X7D
Mg in 2X7E
Mg in 2X7C
Mg in 2X6S
Mg in 2X6U
Mg in 2X7A
Mg in 2X6V
Mg in 2X77