Potassium »
PDB 8qrk-8sfz »
8rz6 »
Potassium in PDB 8rz6: Semet Derivative Structure of the Condensation Domain Tombc From the Tomaymycin Non-Ribosomal Peptide Synthetase
Protein crystallography data
The structure of Semet Derivative Structure of the Condensation Domain Tombc From the Tomaymycin Non-Ribosomal Peptide Synthetase, PDB code: 8rz6
was solved by
M.Karanth,
S.Schmelz,
J.Kirkpatrick,
J.Krausze,
A.Scrima,
T.Carlomagno,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.91 / 2.30 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 123.35, 160.87, 100.15, 90, 125.11, 90 |
| R / Rfree (%) | 17.2 / 21.6 |
Potassium Binding Sites:
The binding sites of Potassium atom in the Semet Derivative Structure of the Condensation Domain Tombc From the Tomaymycin Non-Ribosomal Peptide Synthetase
(pdb code 8rz6). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 4 binding sites of Potassium where determined in the Semet Derivative Structure of the Condensation Domain Tombc From the Tomaymycin Non-Ribosomal Peptide Synthetase, PDB code: 8rz6:
Jump to Potassium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Potassium where determined in the Semet Derivative Structure of the Condensation Domain Tombc From the Tomaymycin Non-Ribosomal Peptide Synthetase, PDB code: 8rz6:
Jump to Potassium binding site number: 1; 2; 3; 4;
Potassium binding site 1 out of 4 in 8rz6
Go back to
Potassium binding site 1 out
of 4 in the Semet Derivative Structure of the Condensation Domain Tombc From the Tomaymycin Non-Ribosomal Peptide Synthetase
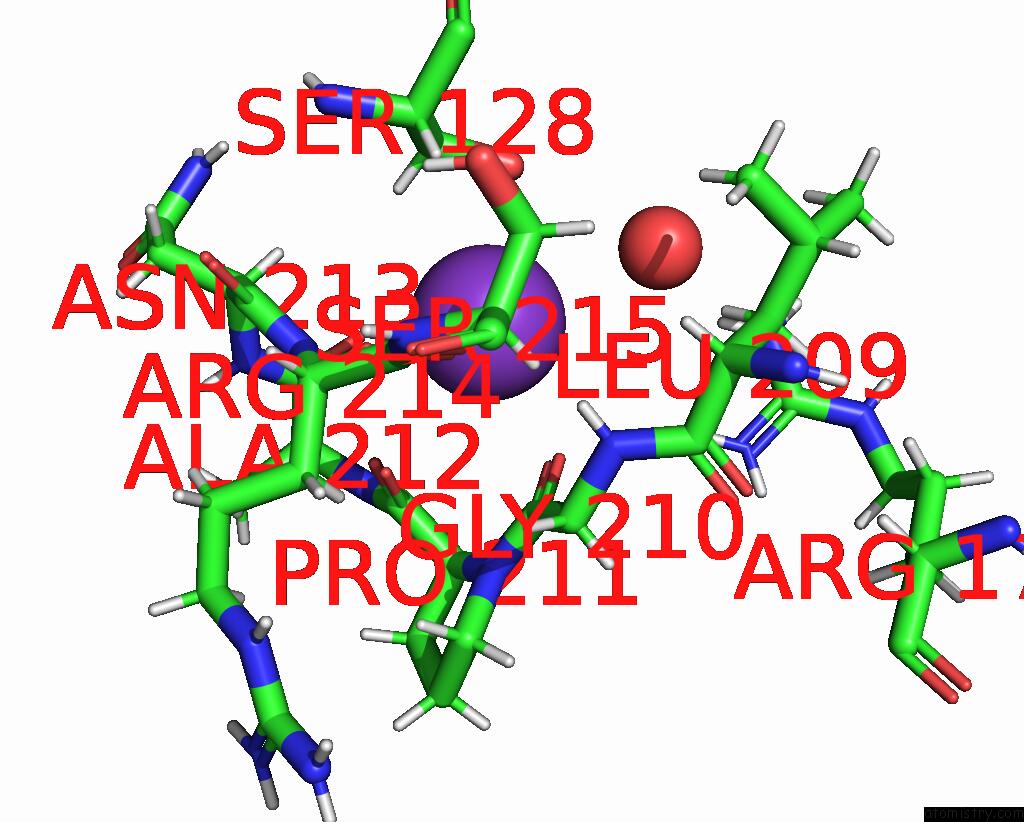
Mono view
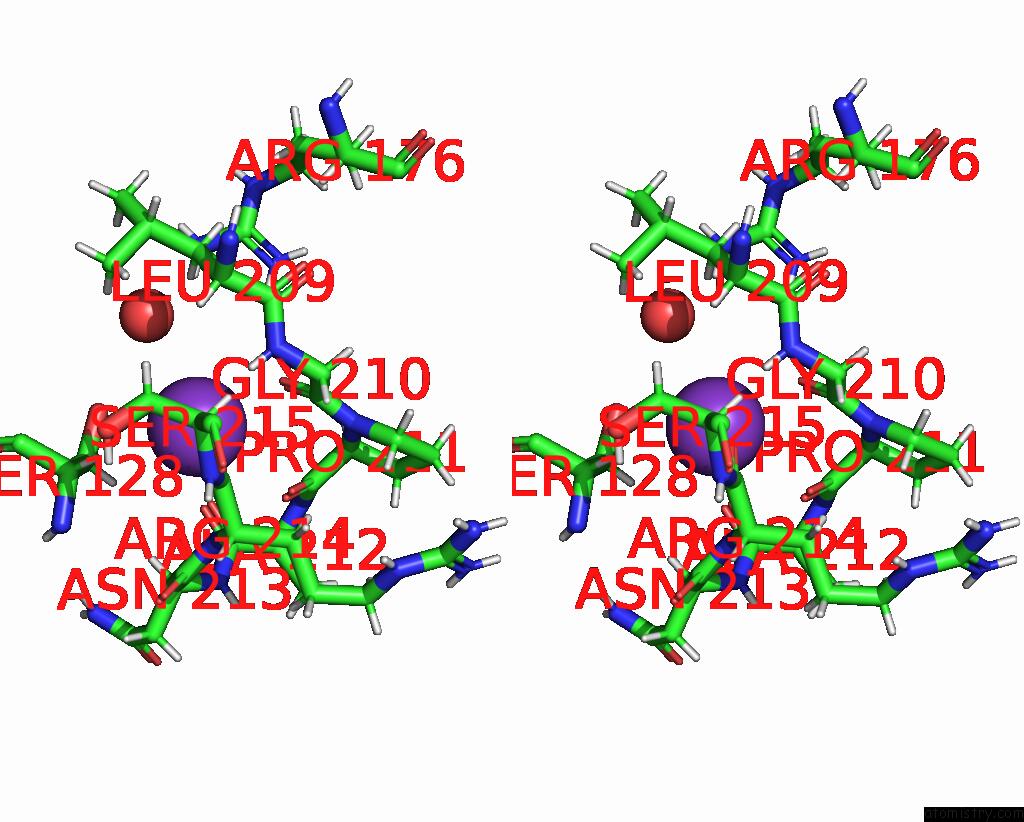
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Semet Derivative Structure of the Condensation Domain Tombc From the Tomaymycin Non-Ribosomal Peptide Synthetase within 5.0Å range:
|
Potassium binding site 2 out of 4 in 8rz6
Go back to
Potassium binding site 2 out
of 4 in the Semet Derivative Structure of the Condensation Domain Tombc From the Tomaymycin Non-Ribosomal Peptide Synthetase
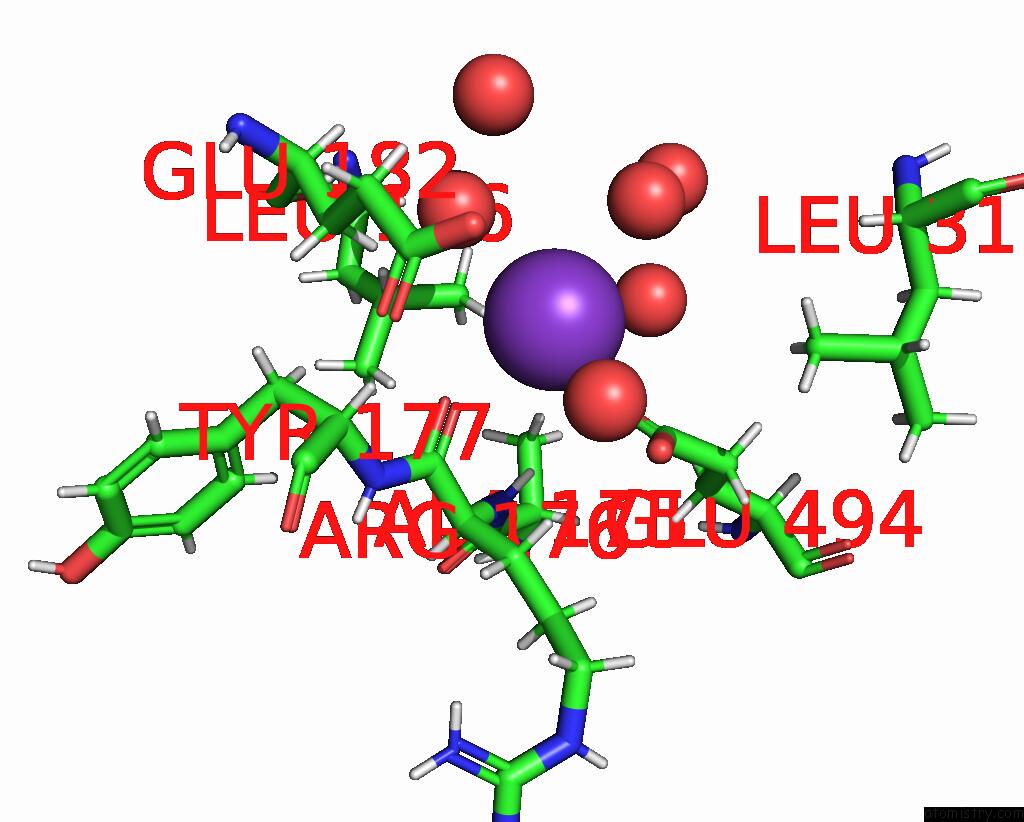
Mono view
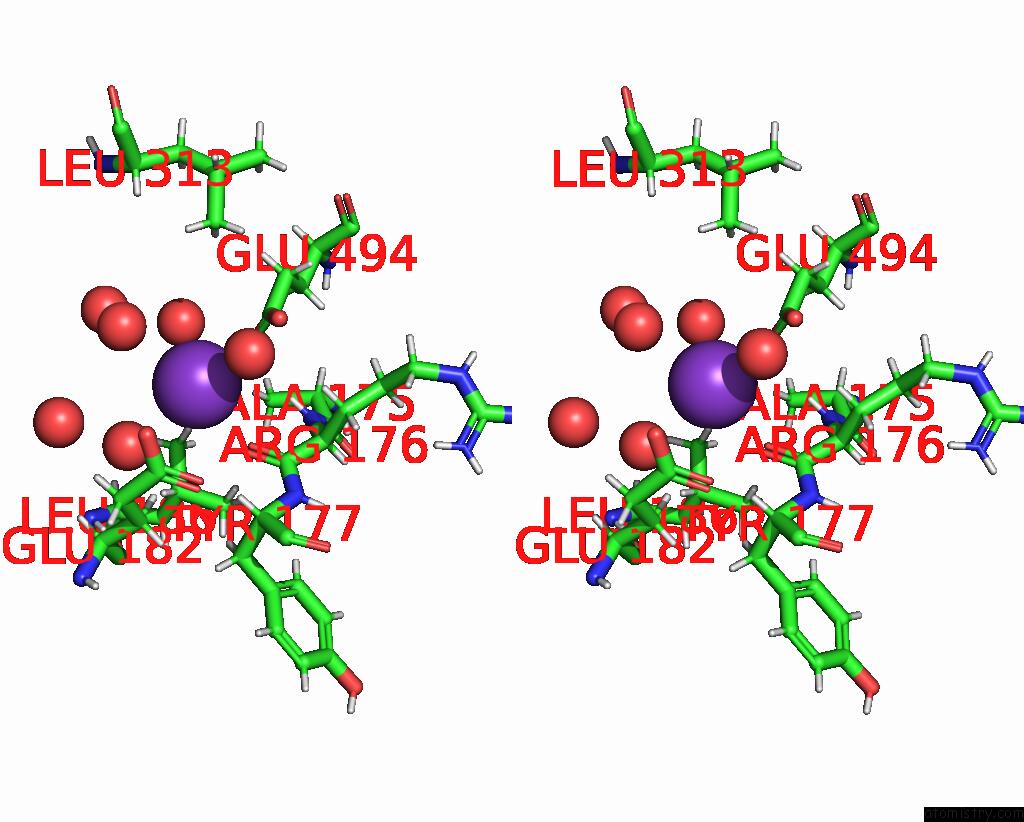
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Semet Derivative Structure of the Condensation Domain Tombc From the Tomaymycin Non-Ribosomal Peptide Synthetase within 5.0Å range:
|
Potassium binding site 3 out of 4 in 8rz6
Go back to
Potassium binding site 3 out
of 4 in the Semet Derivative Structure of the Condensation Domain Tombc From the Tomaymycin Non-Ribosomal Peptide Synthetase
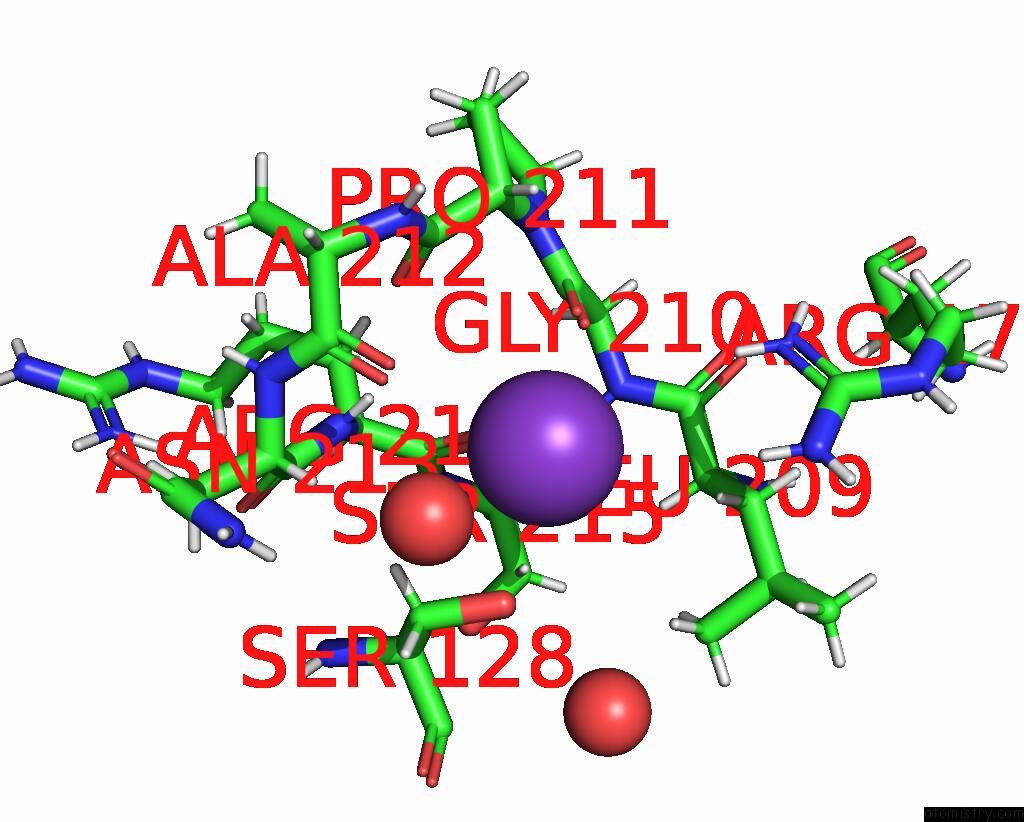
Mono view
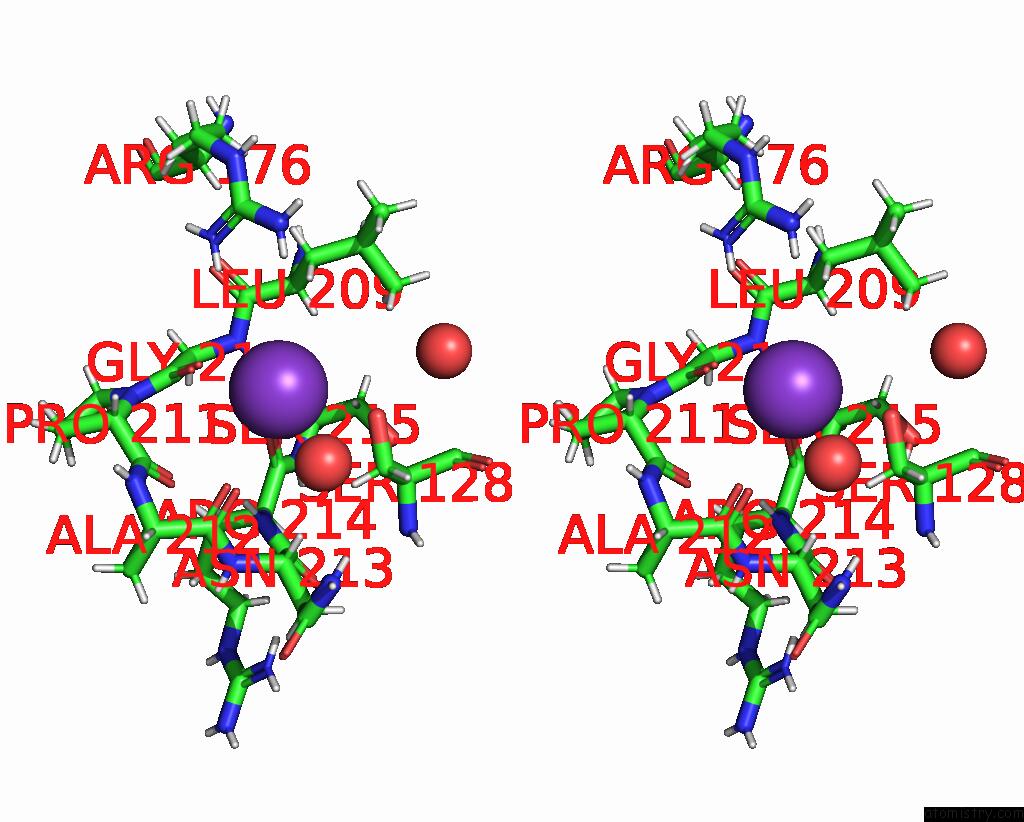
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 3 of Semet Derivative Structure of the Condensation Domain Tombc From the Tomaymycin Non-Ribosomal Peptide Synthetase within 5.0Å range:
|
Potassium binding site 4 out of 4 in 8rz6
Go back to
Potassium binding site 4 out
of 4 in the Semet Derivative Structure of the Condensation Domain Tombc From the Tomaymycin Non-Ribosomal Peptide Synthetase
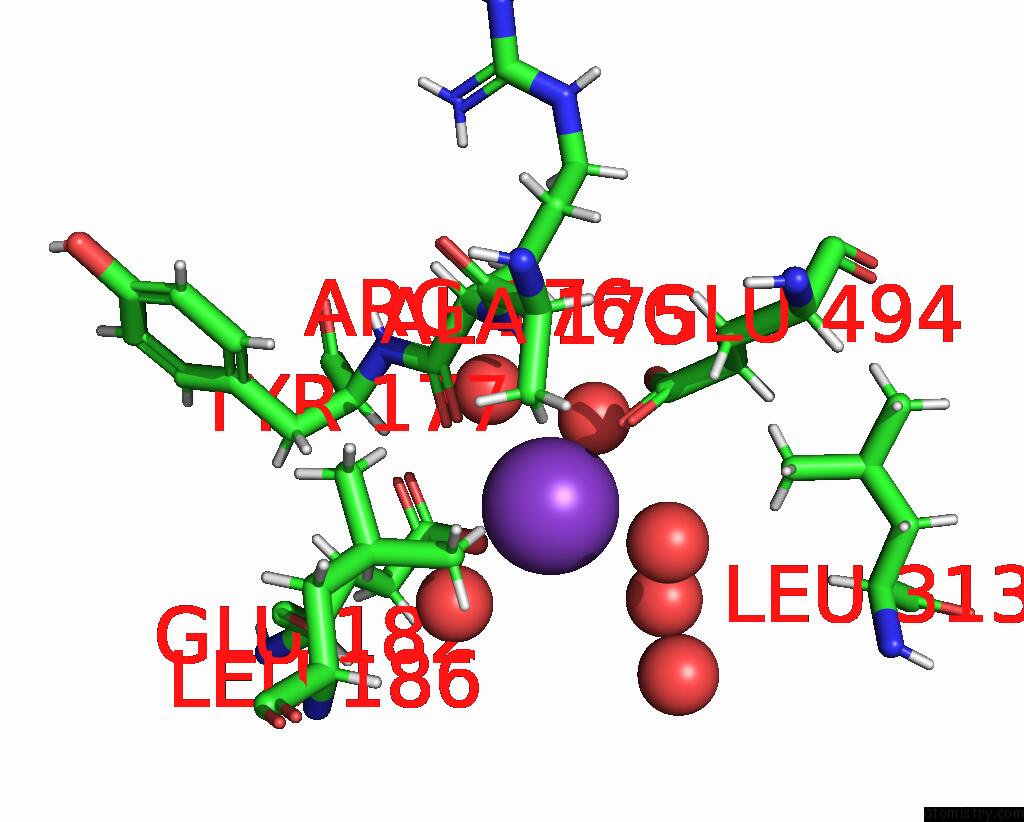
Mono view
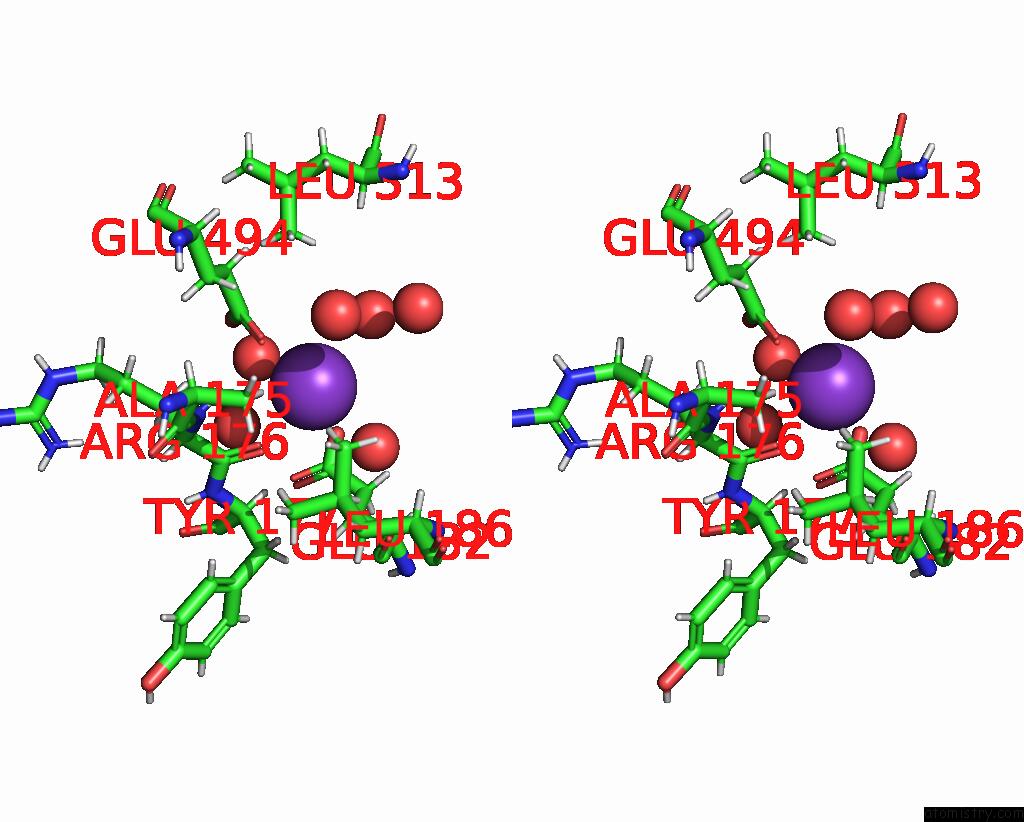
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 4 of Semet Derivative Structure of the Condensation Domain Tombc From the Tomaymycin Non-Ribosomal Peptide Synthetase within 5.0Å range:
|
Reference:
M.N.Karanth,
J.P.Kirkpatrick,
J.Krausze,
S.Schmelz,
A.Scrima,
T.Carlomagno.
The Specificity of Intermodular Recognition in A Prototypical Nonribosomal Peptide Synthetase Depends on An Adaptor Domain Sci Adv V. 10 2024.
ISSN: ESSN 2375-2548
DOI: 10.1126/SCIADV.ADM9404
Page generated: Sat Aug 9 17:43:09 2025
ISSN: ESSN 2375-2548
DOI: 10.1126/SCIADV.ADM9404
Last articles
Mg in 2X0QMg in 2X03
Mg in 2WZG
Mg in 2WZD
Mg in 2WZC
Mg in 2WZB
Mg in 2WZ8
Mg in 2WX5
Mg in 2WWR
Mg in 2WW8