Potassium »
PDB 8qrk-8sfz »
8qz3 »
Potassium in PDB 8qz3: Crystal Structure of Human Two Pore Domain Potassium Ion Channel Trek- 2 (K2P10.1) in Complex with An Activatory Nanobody (NB67)
Protein crystallography data
The structure of Crystal Structure of Human Two Pore Domain Potassium Ion Channel Trek- 2 (K2P10.1) in Complex with An Activatory Nanobody (NB67), PDB code: 8qz3
was solved by
A.Baronina,
A.C.W.Pike,
K.E.J.Rodstrom,
J.Ang,
S.R.Bushell,
R.Chalk,
S.M.M.Mukhopadhyay,
E.Pardon,
C.H.Arrowsmith,
A.M.Edwards,
C.Bountra,
N.A.Burgess-Brown,
S.J.Tucker,
J.Steyaert,
E.P.Carpenter,
Structuralgenomics Consortium (Sgc),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 58.81 / 2.40 |
| Space group | P 2 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 51.264, 102.1, 215.817, 90, 90, 90 |
| R / Rfree (%) | 27.8 / 29.1 |
Potassium Binding Sites:
The binding sites of Potassium atom in the Crystal Structure of Human Two Pore Domain Potassium Ion Channel Trek- 2 (K2P10.1) in Complex with An Activatory Nanobody (NB67)
(pdb code 8qz3). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 5 binding sites of Potassium where determined in the Crystal Structure of Human Two Pore Domain Potassium Ion Channel Trek- 2 (K2P10.1) in Complex with An Activatory Nanobody (NB67), PDB code: 8qz3:
Jump to Potassium binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Potassium where determined in the Crystal Structure of Human Two Pore Domain Potassium Ion Channel Trek- 2 (K2P10.1) in Complex with An Activatory Nanobody (NB67), PDB code: 8qz3:
Jump to Potassium binding site number: 1; 2; 3; 4; 5;
Potassium binding site 1 out of 5 in 8qz3
Go back to
Potassium binding site 1 out
of 5 in the Crystal Structure of Human Two Pore Domain Potassium Ion Channel Trek- 2 (K2P10.1) in Complex with An Activatory Nanobody (NB67)
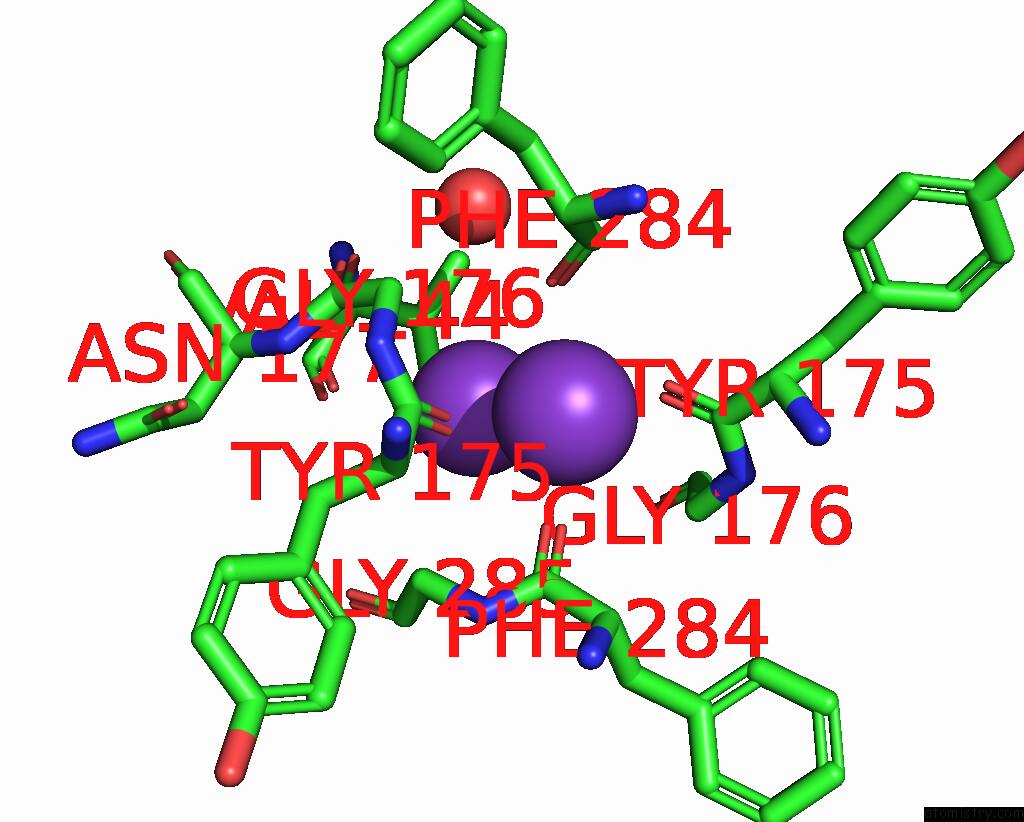
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Crystal Structure of Human Two Pore Domain Potassium Ion Channel Trek- 2 (K2P10.1) in Complex with An Activatory Nanobody (NB67) within 5.0Å range:
|
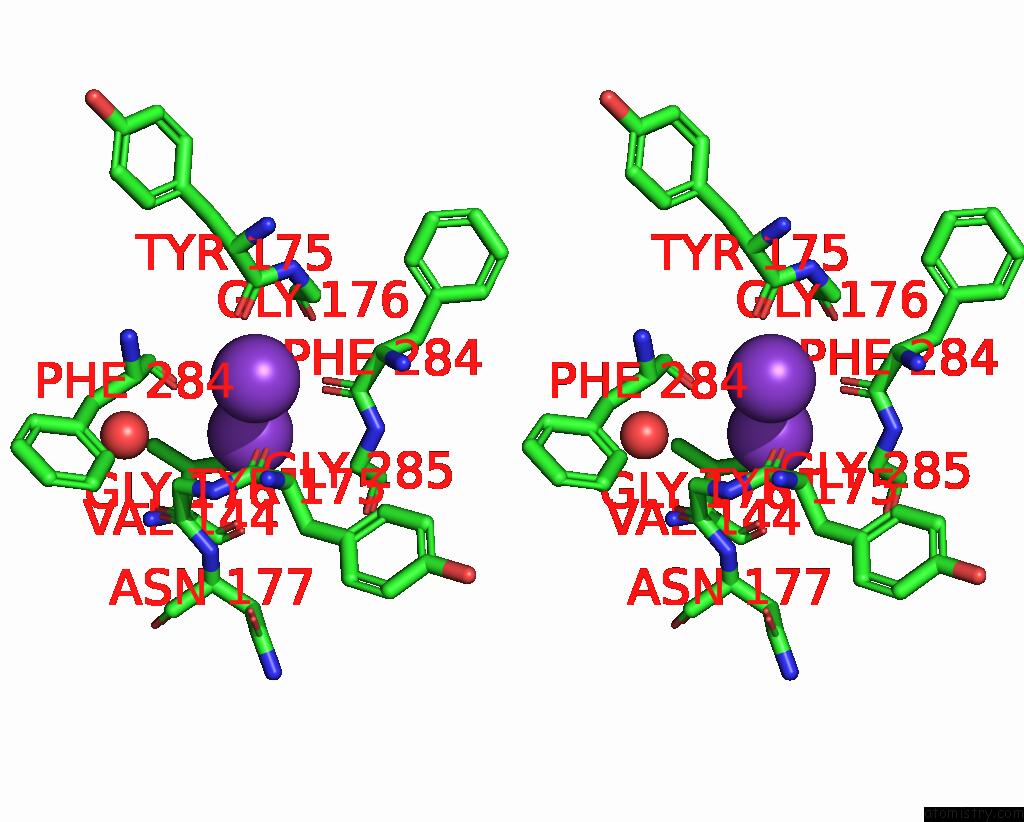
Potassium binding site 2 out of 5 in 8qz3
Go back to
Potassium binding site 2 out
of 5 in the Crystal Structure of Human Two Pore Domain Potassium Ion Channel Trek- 2 (K2P10.1) in Complex with An Activatory Nanobody (NB67)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Crystal Structure of Human Two Pore Domain Potassium Ion Channel Trek- 2 (K2P10.1) in Complex with An Activatory Nanobody (NB67) within 5.0Å range:
|
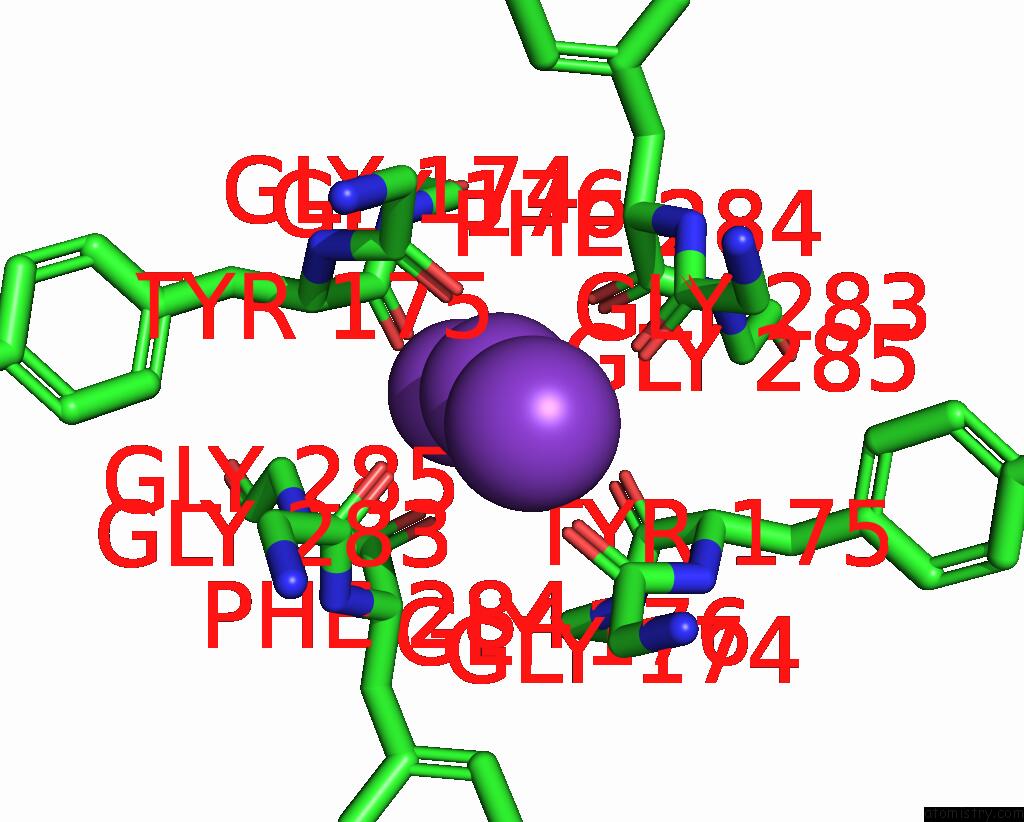
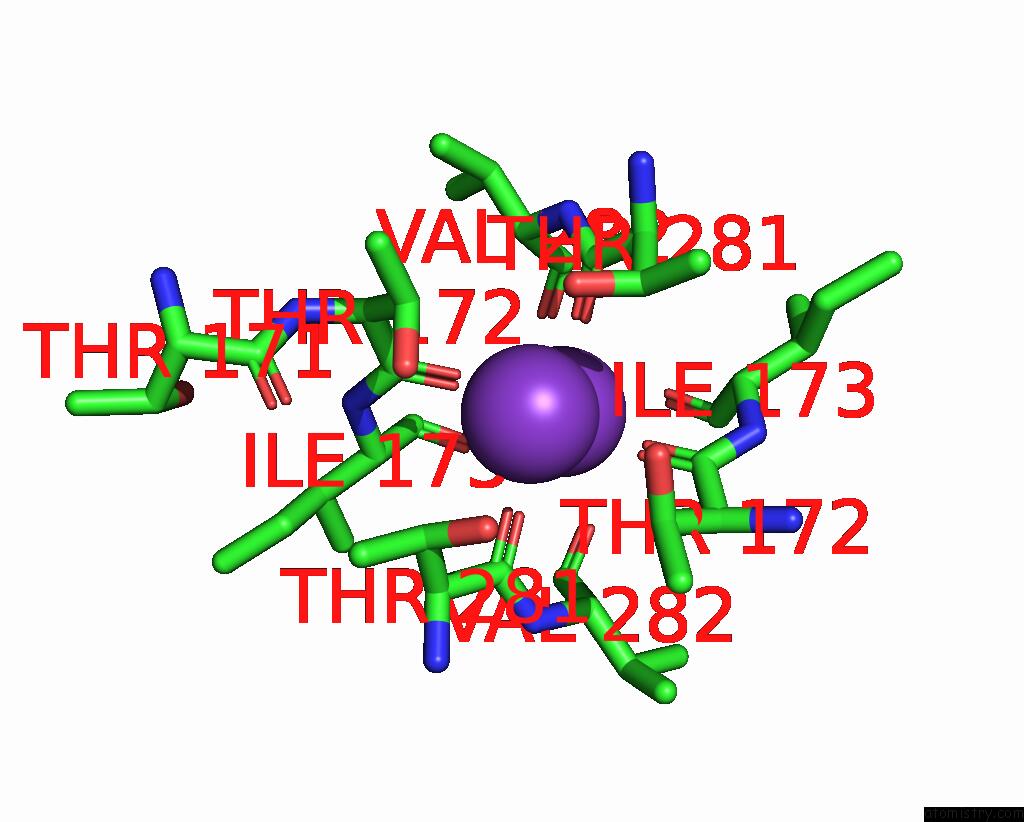
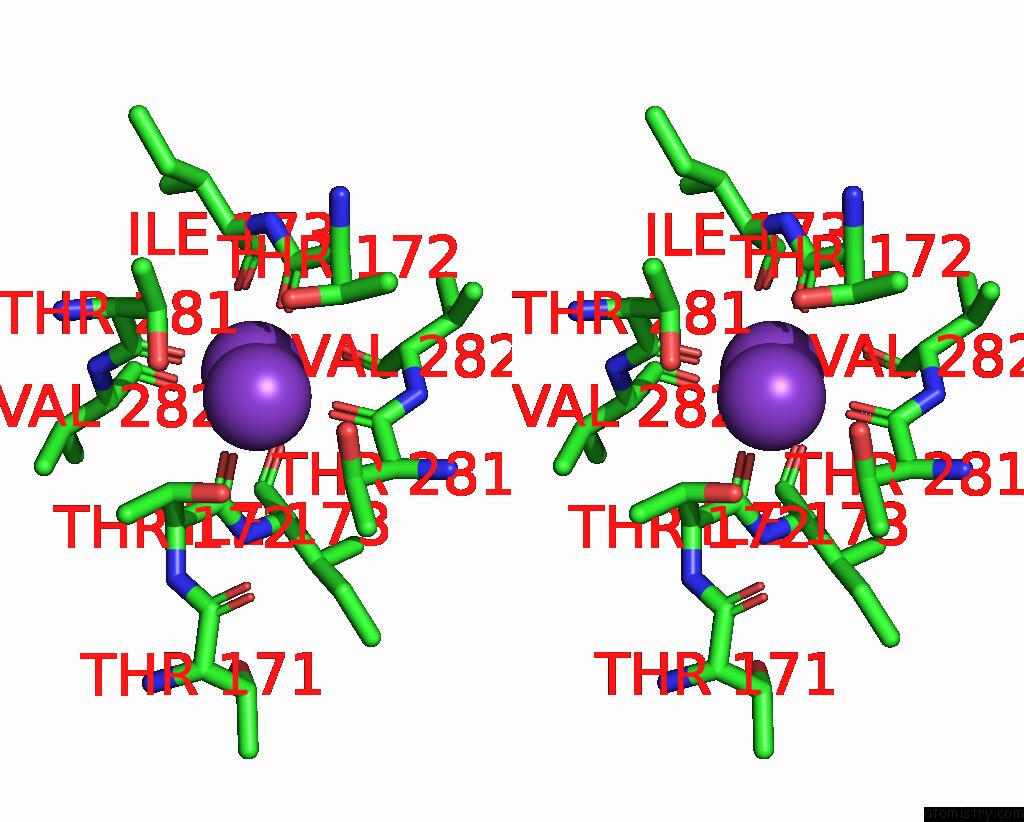
Potassium binding site 3 out of 5 in 8qz3
Go back to
Potassium binding site 3 out
of 5 in the Crystal Structure of Human Two Pore Domain Potassium Ion Channel Trek- 2 (K2P10.1) in Complex with An Activatory Nanobody (NB67)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 3 of Crystal Structure of Human Two Pore Domain Potassium Ion Channel Trek- 2 (K2P10.1) in Complex with An Activatory Nanobody (NB67) within 5.0Å range:
|
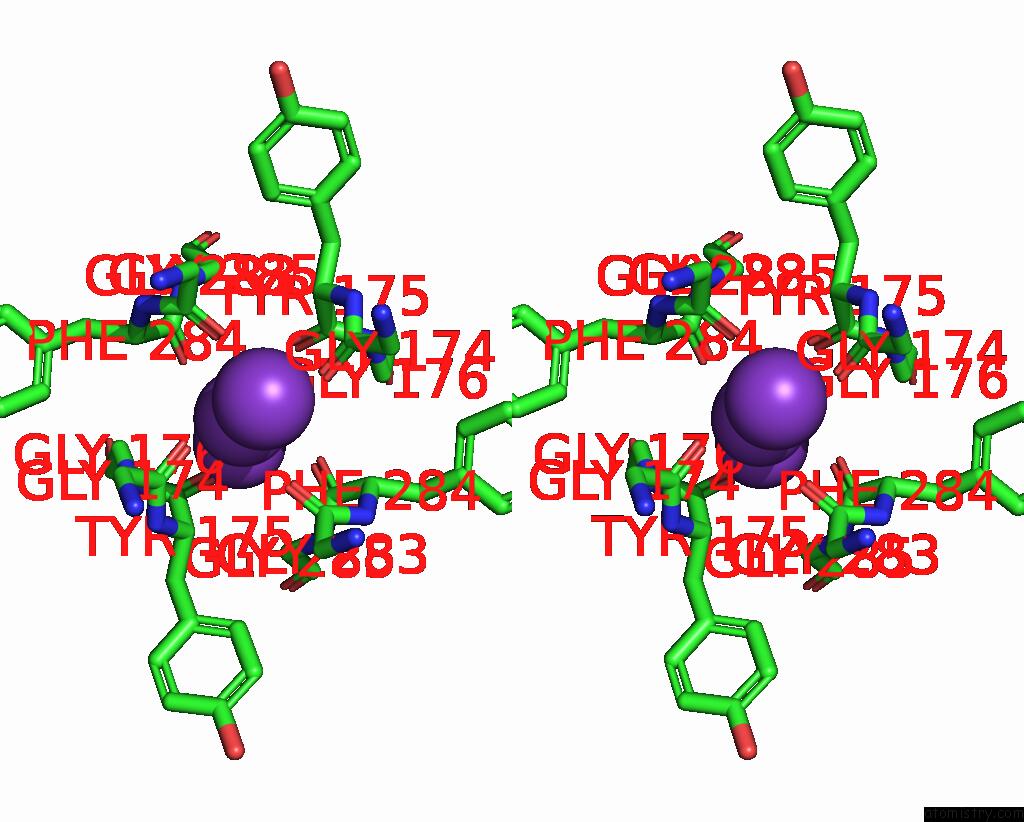
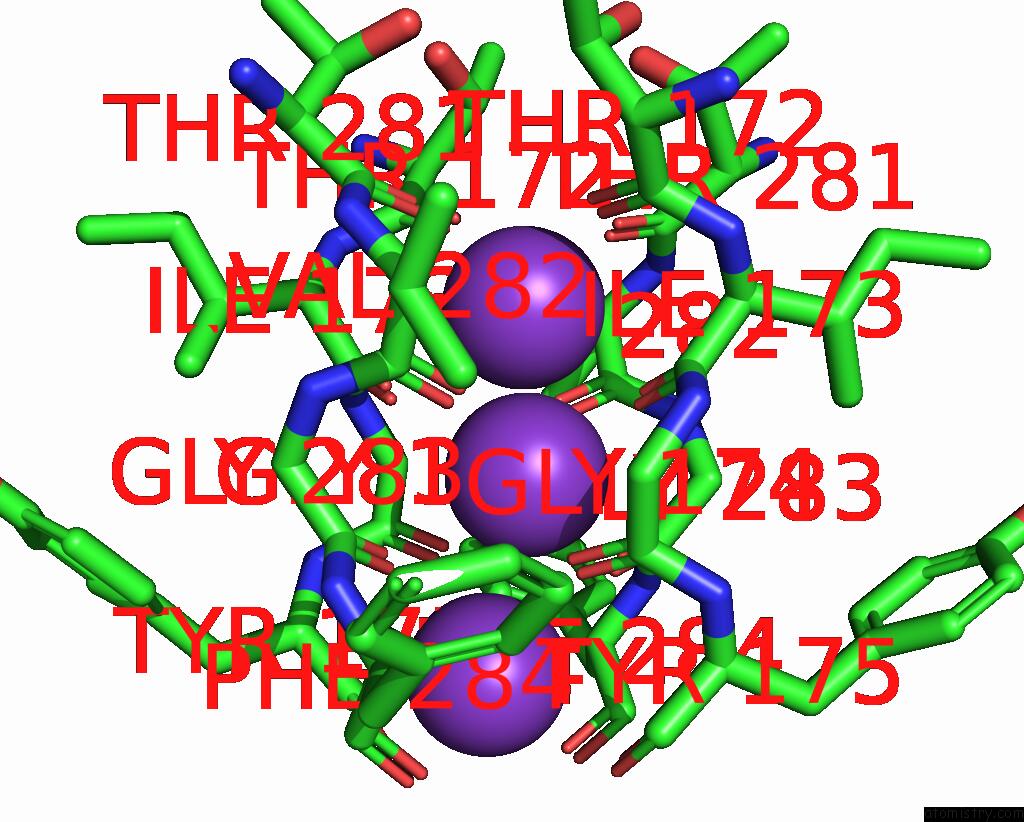
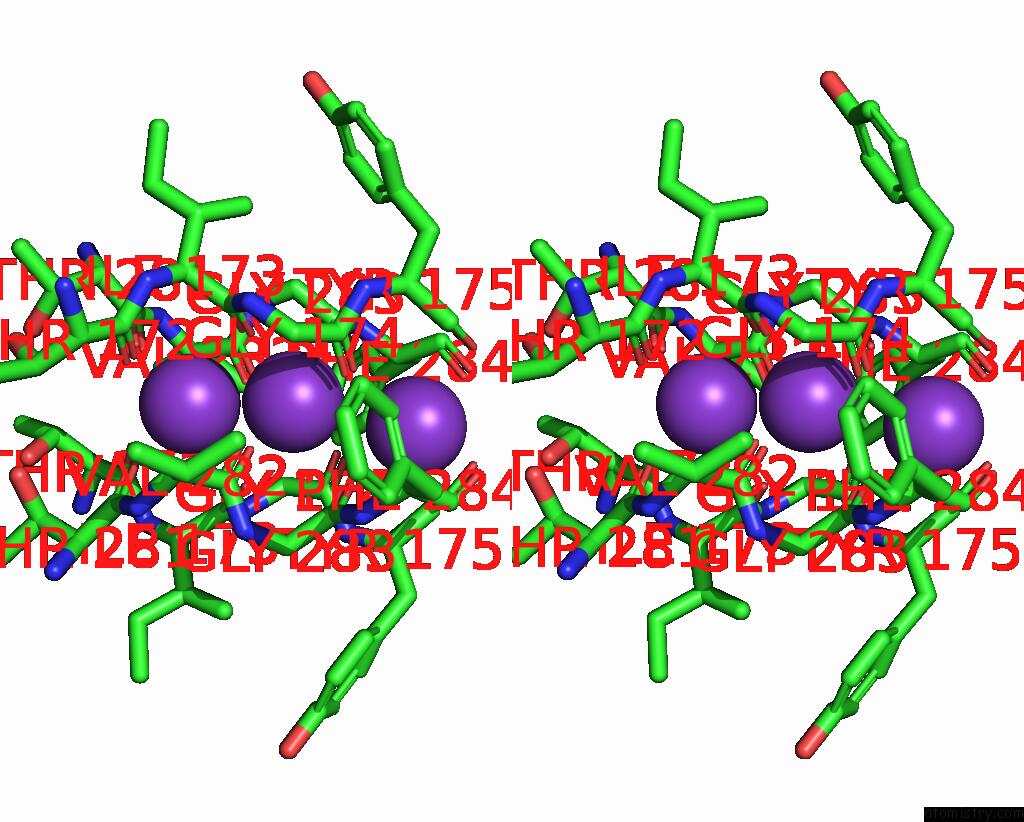
Potassium binding site 4 out of 5 in 8qz3
Go back to
Potassium binding site 4 out
of 5 in the Crystal Structure of Human Two Pore Domain Potassium Ion Channel Trek- 2 (K2P10.1) in Complex with An Activatory Nanobody (NB67)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 4 of Crystal Structure of Human Two Pore Domain Potassium Ion Channel Trek- 2 (K2P10.1) in Complex with An Activatory Nanobody (NB67) within 5.0Å range:
|
Potassium binding site 5 out of 5 in 8qz3
Go back to
Potassium binding site 5 out
of 5 in the Crystal Structure of Human Two Pore Domain Potassium Ion Channel Trek- 2 (K2P10.1) in Complex with An Activatory Nanobody (NB67)
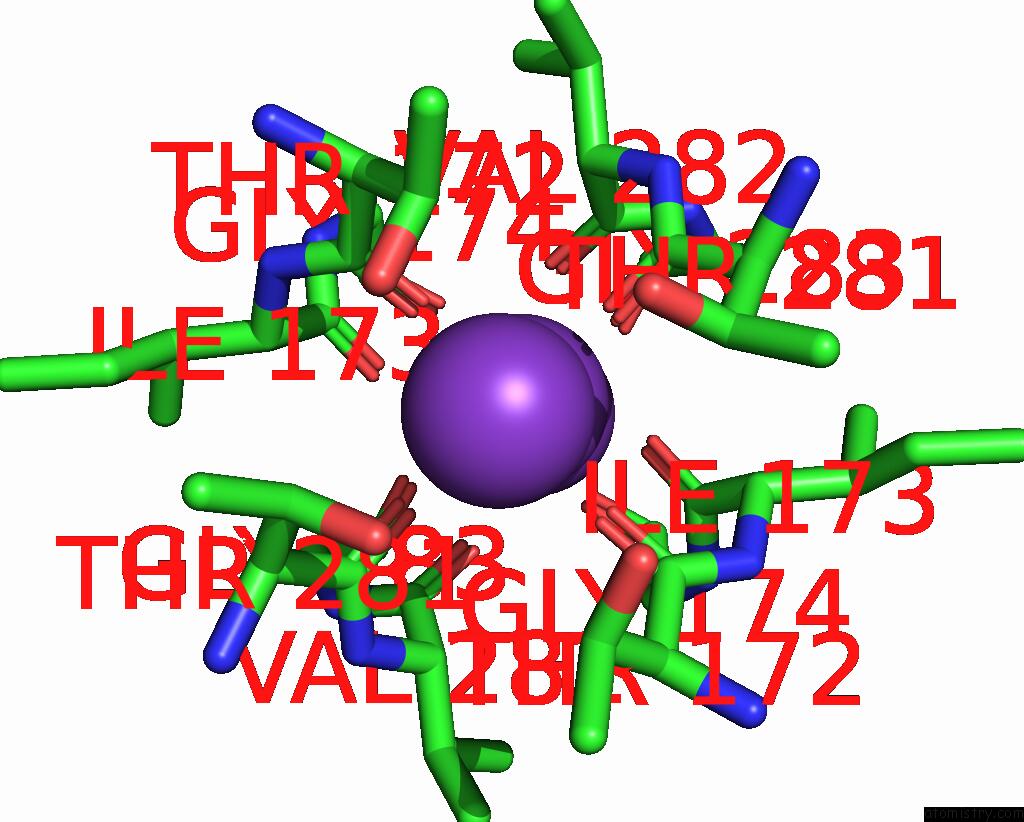
Mono view
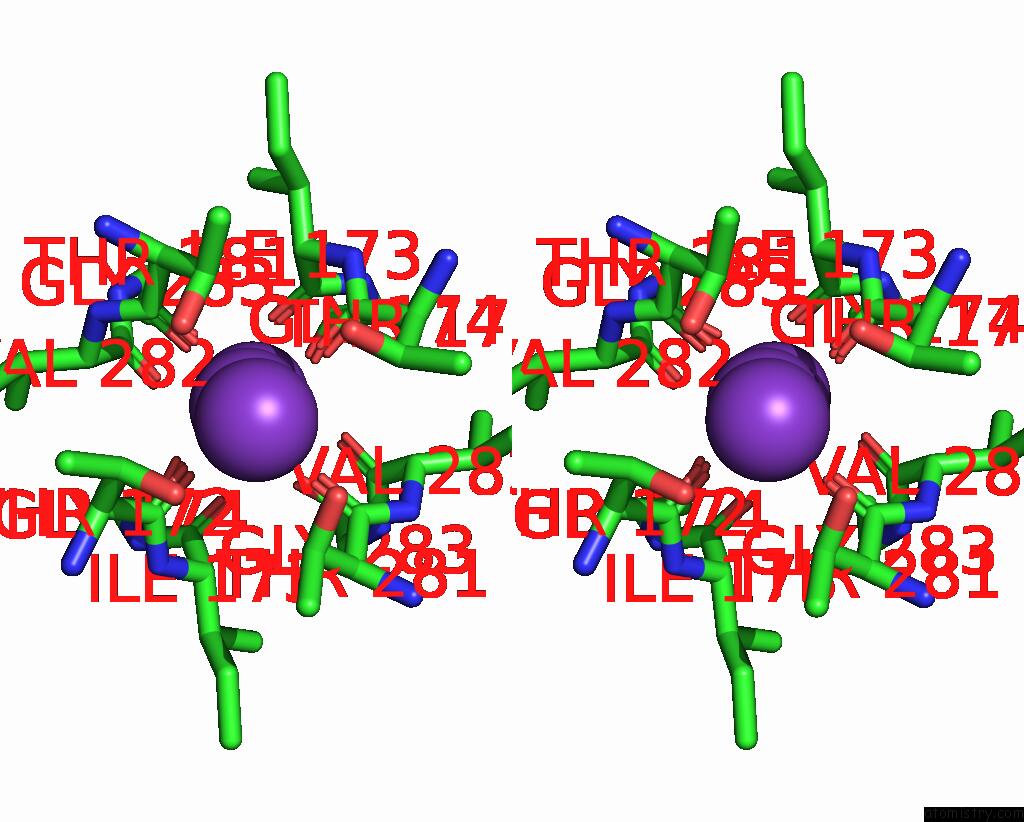
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 5 of Crystal Structure of Human Two Pore Domain Potassium Ion Channel Trek- 2 (K2P10.1) in Complex with An Activatory Nanobody (NB67) within 5.0Å range:
|
Reference:
K.E.J.Rodstrom,
A.Cloake,
J.Sormann,
A.Baronina,
K.H.M.Smith,
A.C.W.Pike,
J.Ang,
P.Proks,
M.Schewe,
I.Holland-Kaye,
S.R.Bushell,
J.Elliott,
E.Pardon,
T.Baukrowitz,
R.J.Owens,
S.Newstead,
J.Steyaert,
E.P.Carpenter,
S.J.Tucker.
Extracellular Modulation of Trek-2 Activity with Nanobodies Provides Insight Into the Mechanisms of K2P Channel Regulation. Nat Commun V. 15 4173 2024.
ISSN: ESSN 2041-1723
PubMed: 38755204
DOI: 10.1038/S41467-024-48536-2
Page generated: Sat Aug 9 17:38:17 2025
ISSN: ESSN 2041-1723
PubMed: 38755204
DOI: 10.1038/S41467-024-48536-2
Last articles
Mg in 1JBZMg in 1JBW
Mg in 1JBV
Mg in 1JBK
Mg in 1JAX
Mg in 1JAH
Mg in 1J97
Mg in 1J9J
Mg in 1J8L
Mg in 1J7U