Potassium »
PDB 8qrk-8sfz »
8qxu »
Potassium in PDB 8qxu: In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
Enzymatic activity of In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
All present enzymatic activity of In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography:
5.6.1.7;
5.6.1.7;
Other elements in 8qxu:
The structure of In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography also contains other interesting chemical elements:
| Magnesium | (Mg) | 14 atoms |
Potassium Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 14;Binding sites:
The binding sites of Potassium atom in the In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography (pdb code 8qxu). This binding sites where shown within 5.0 Angstroms radius around Potassium atom.In total 14 binding sites of Potassium where determined in the In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography, PDB code: 8qxu:
Jump to Potassium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Potassium binding site 1 out of 14 in 8qxu
Go back to
Potassium binding site 1 out
of 14 in the In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
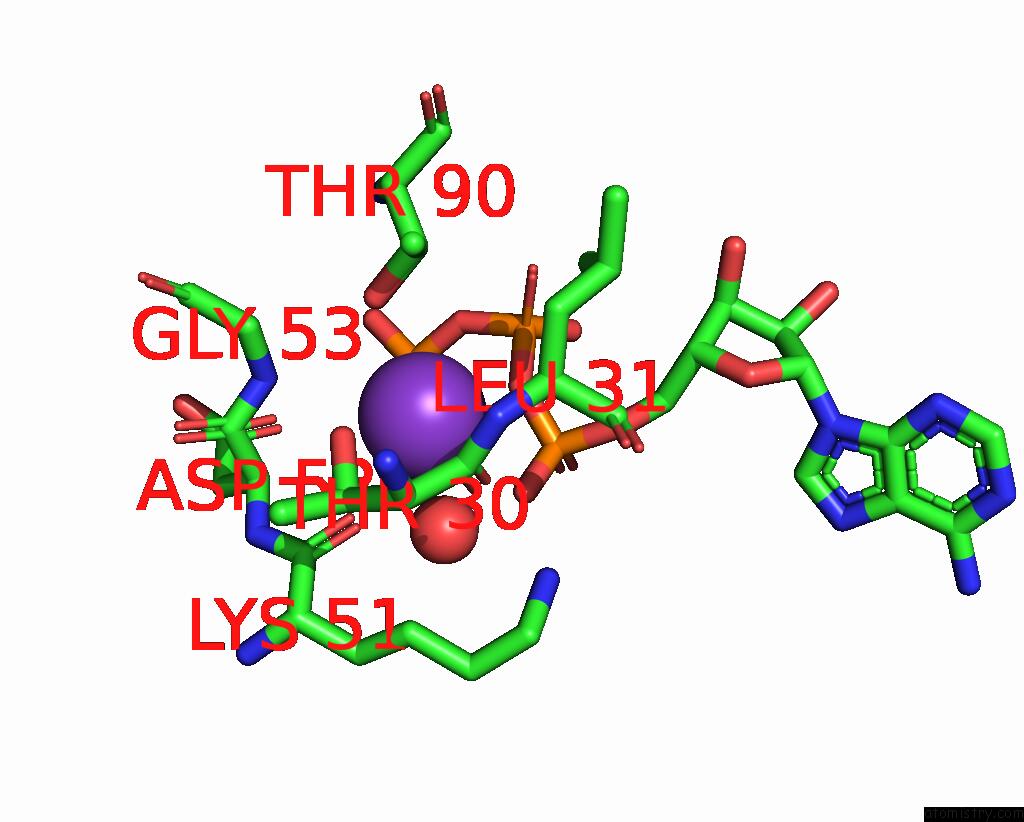
Mono view
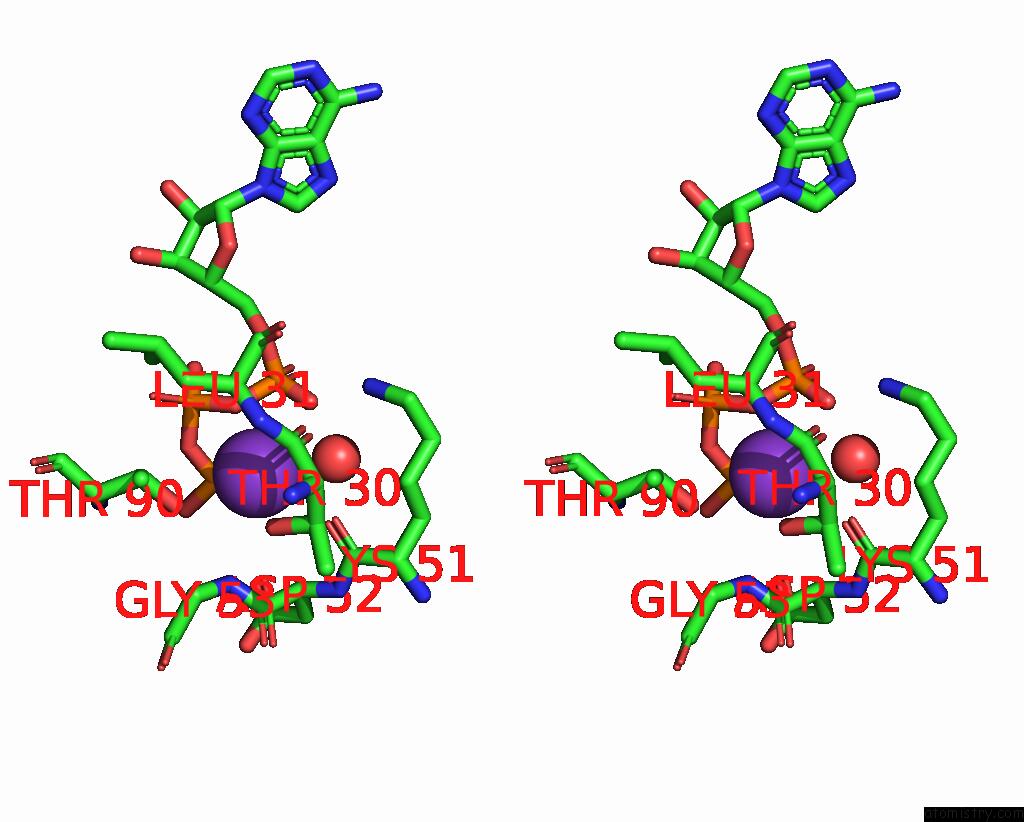
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography within 5.0Å range:
|
Potassium binding site 2 out of 14 in 8qxu
Go back to
Potassium binding site 2 out
of 14 in the In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
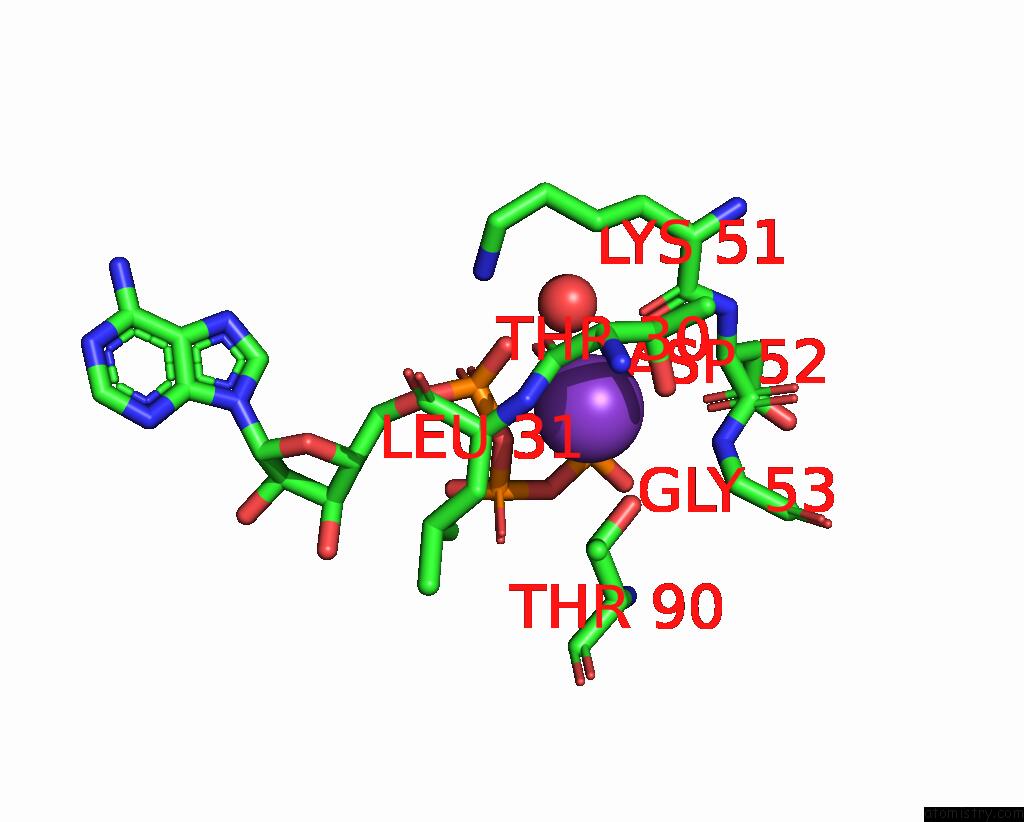
Mono view
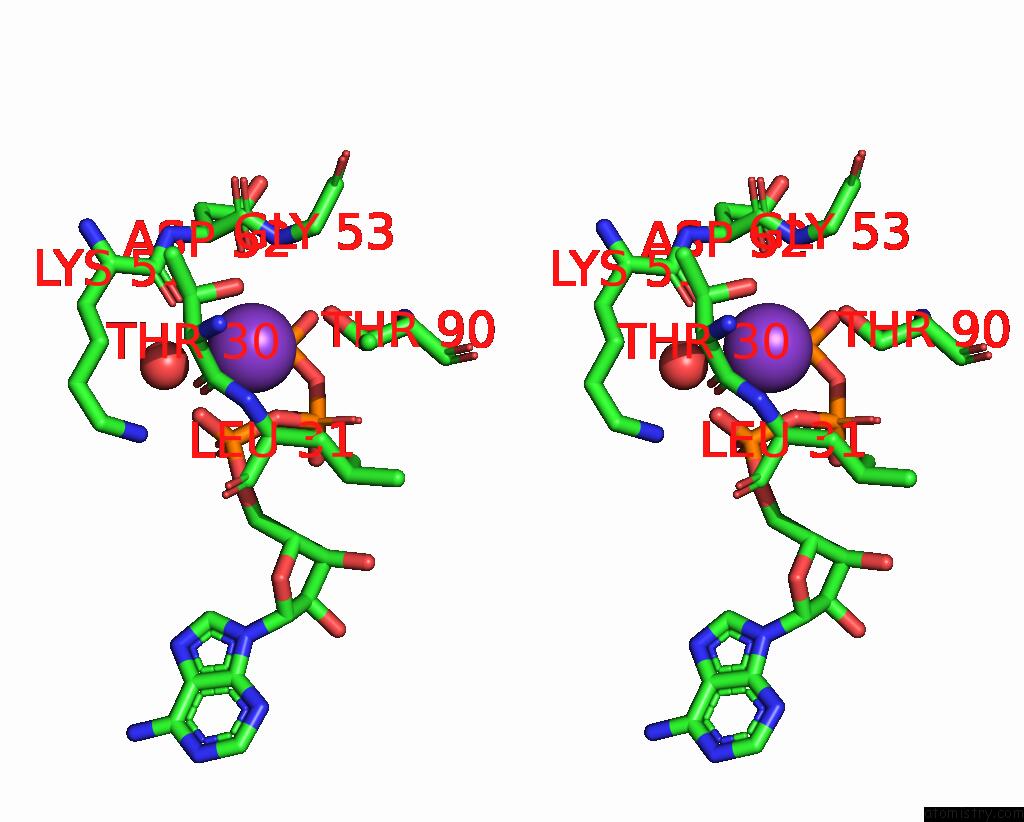
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography within 5.0Å range:
|
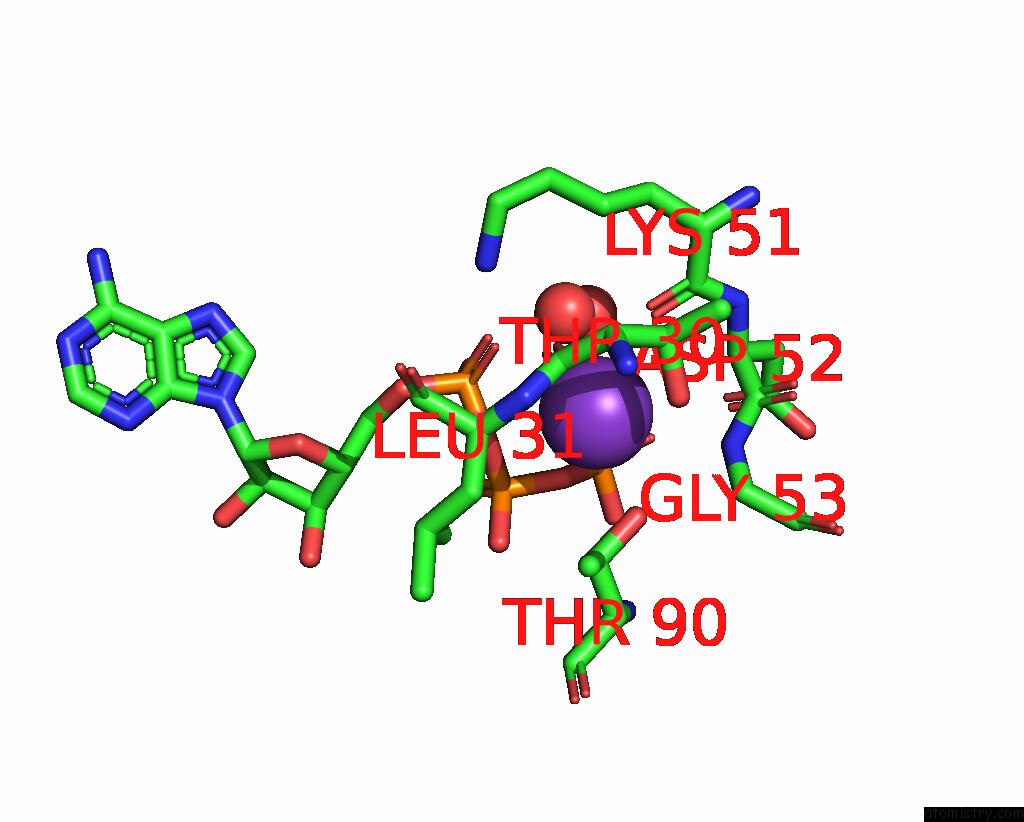
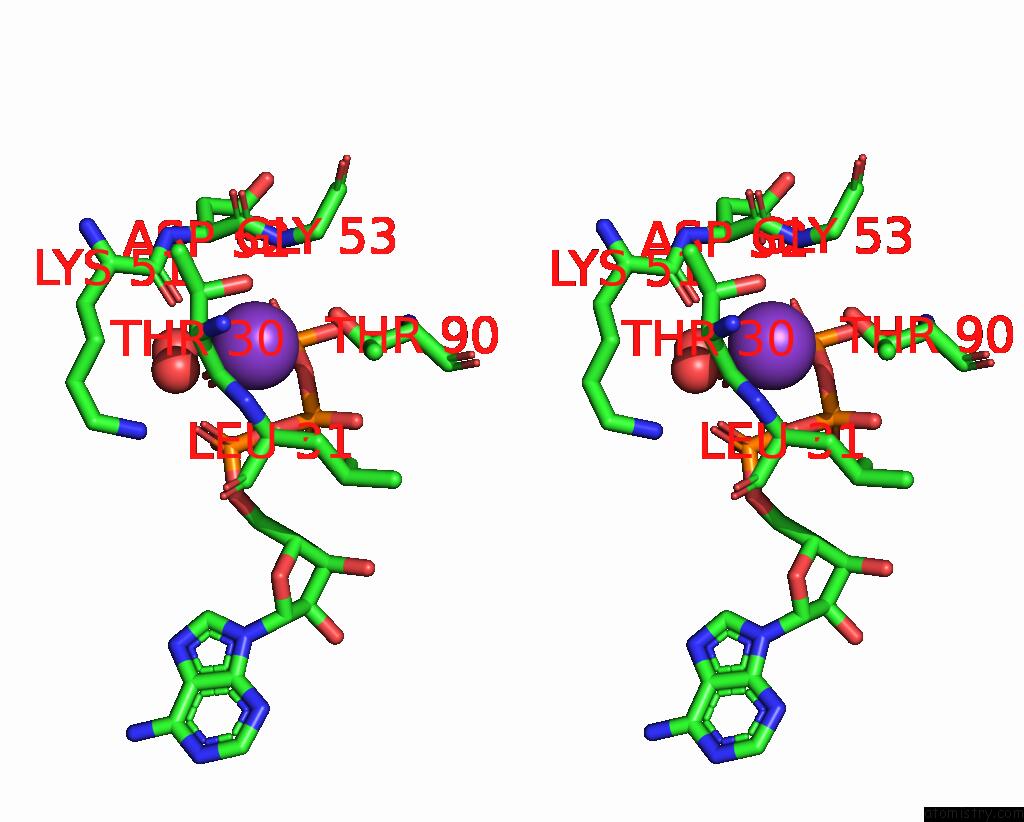
Potassium binding site 3 out of 14 in 8qxu
Go back to
Potassium binding site 3 out
of 14 in the In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 3 of In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography within 5.0Å range:
|
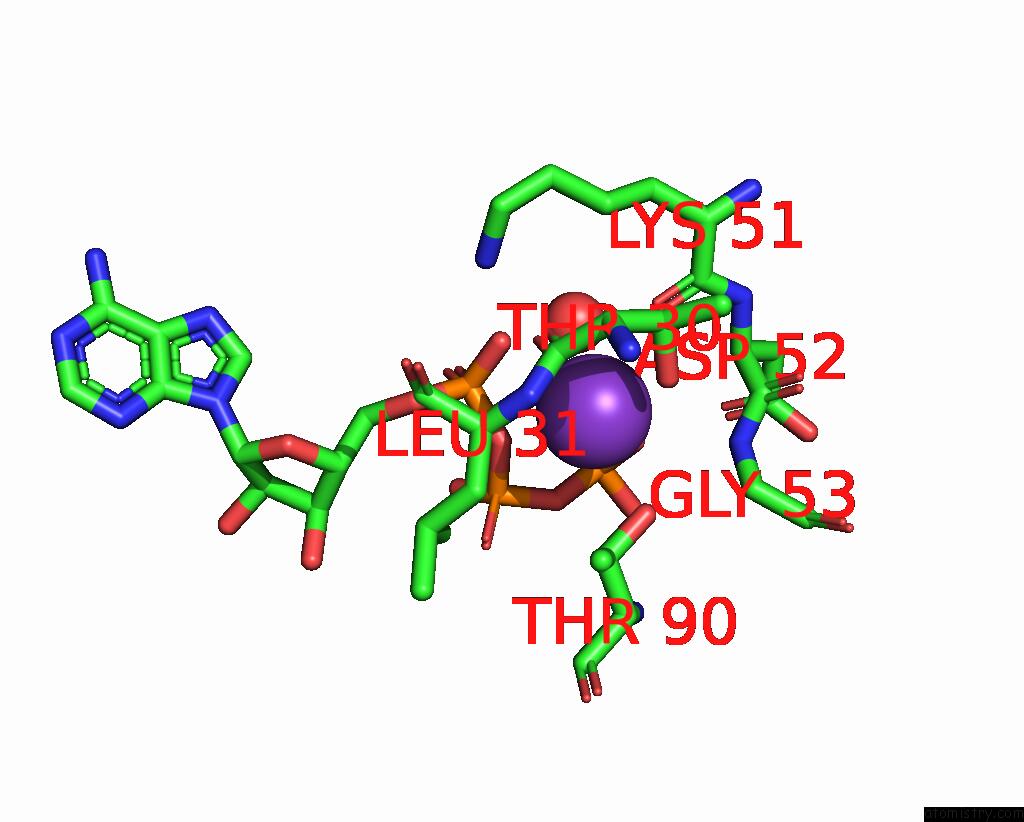
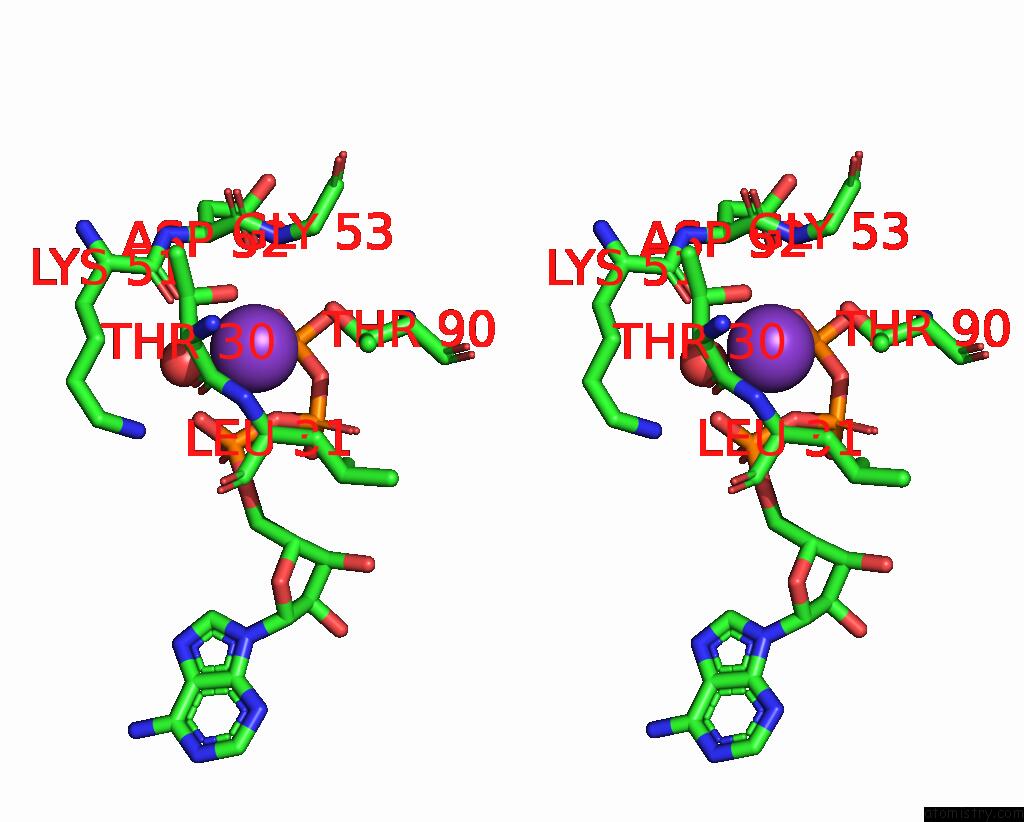
Potassium binding site 4 out of 14 in 8qxu
Go back to
Potassium binding site 4 out
of 14 in the In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 4 of In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography within 5.0Å range:
|
Potassium binding site 5 out of 14 in 8qxu
Go back to
Potassium binding site 5 out
of 14 in the In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
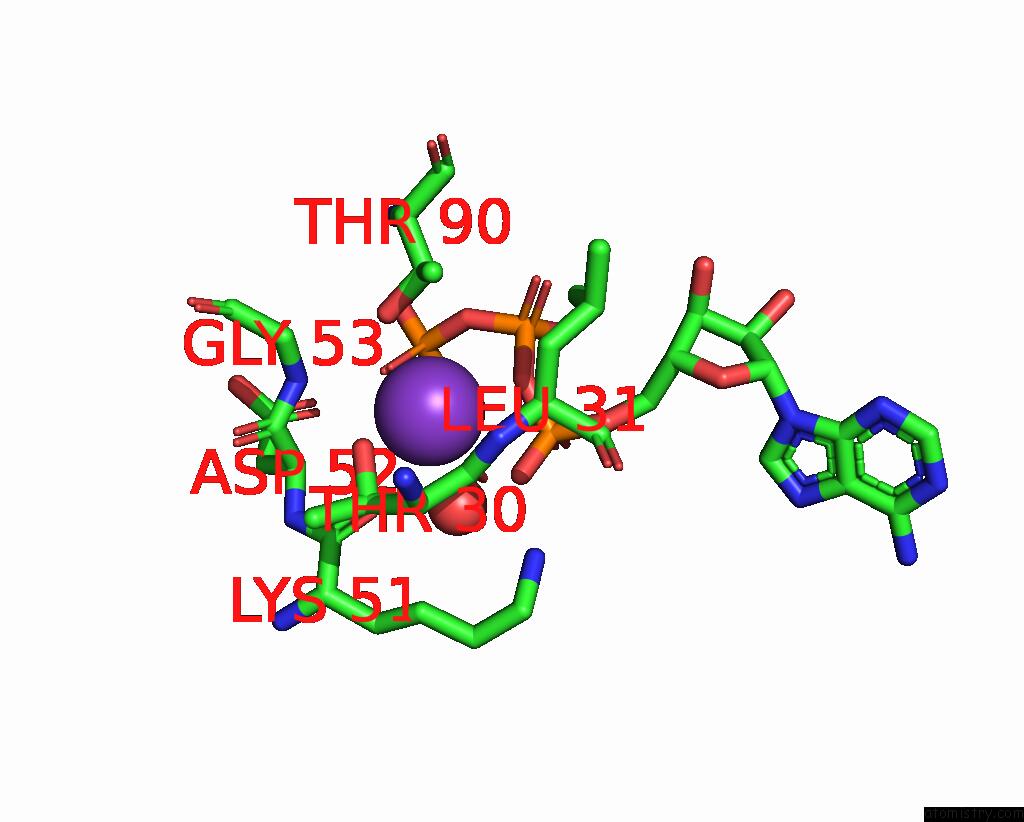
Mono view
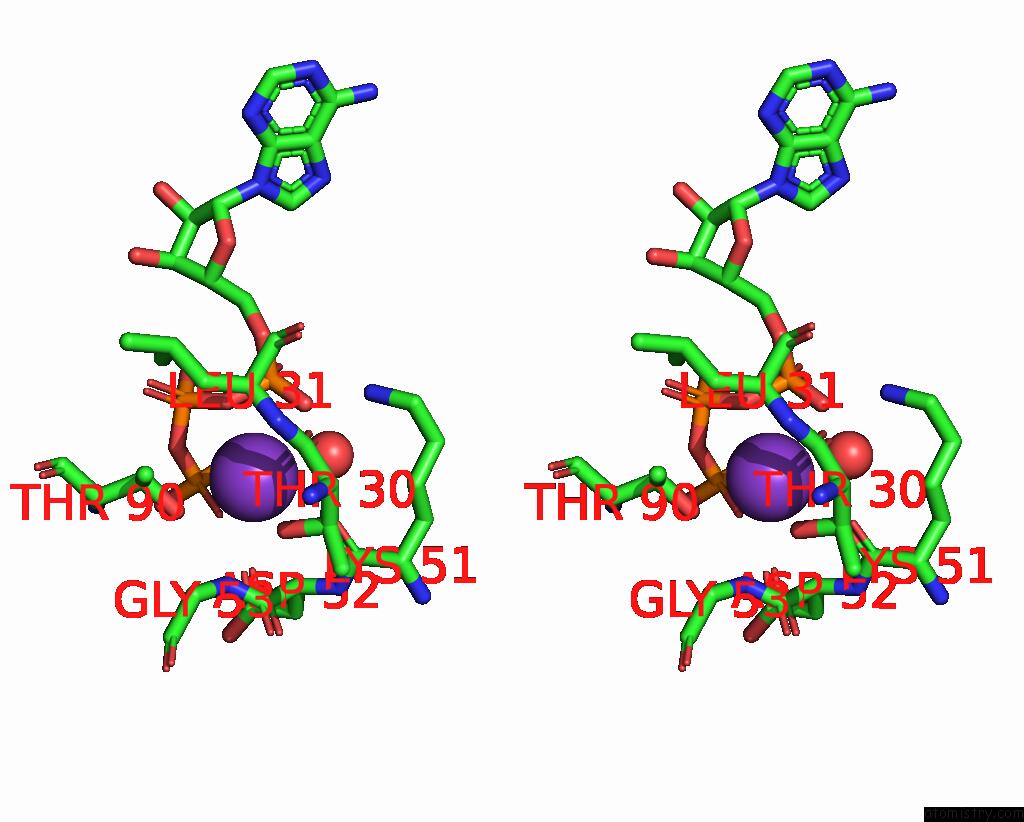
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 5 of In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography within 5.0Å range:
|
Potassium binding site 6 out of 14 in 8qxu
Go back to
Potassium binding site 6 out
of 14 in the In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
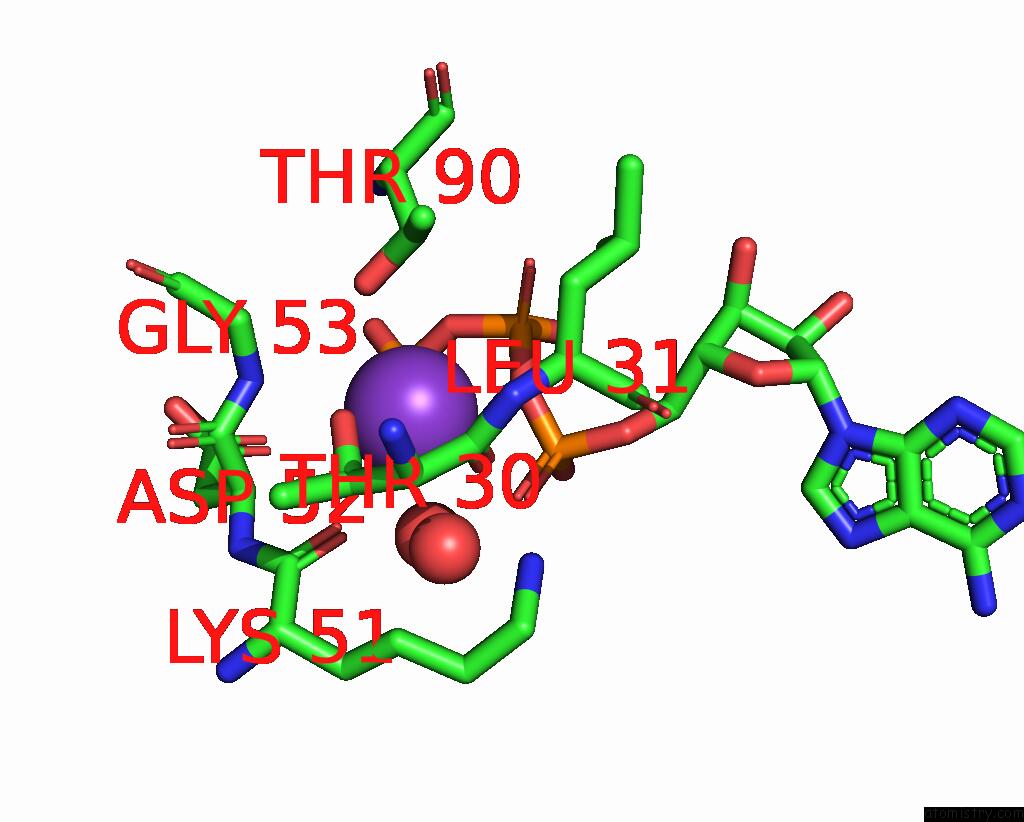
Mono view
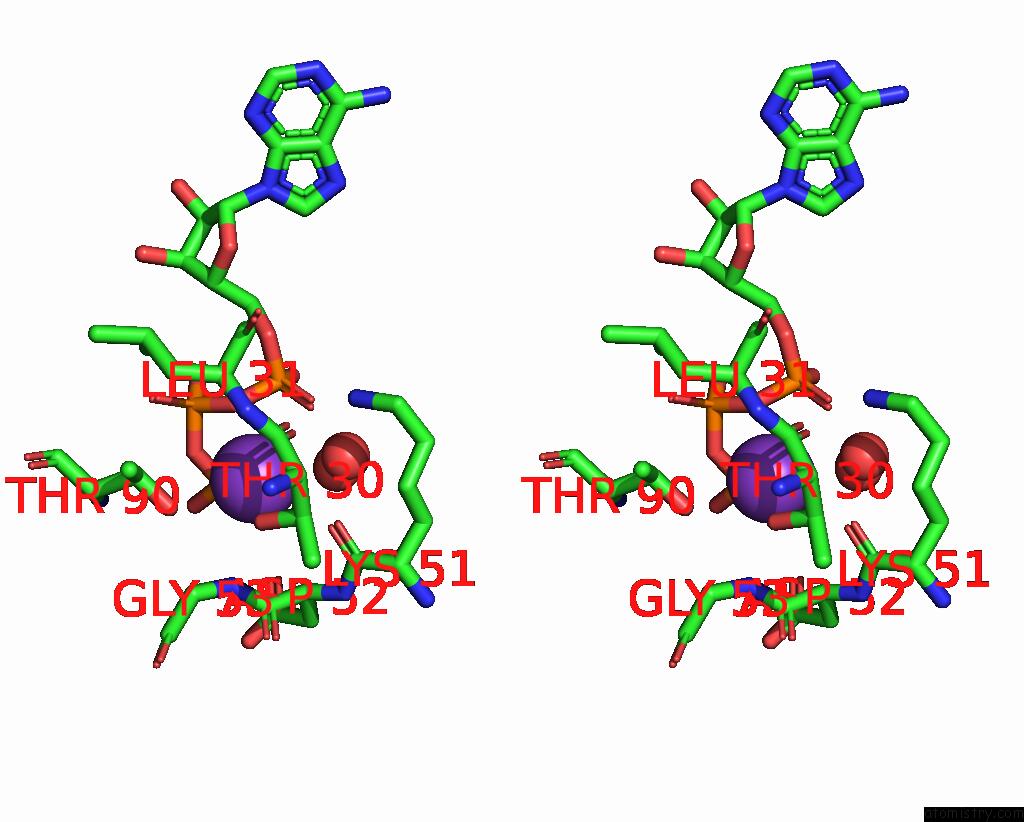
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 6 of In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography within 5.0Å range:
|
Potassium binding site 7 out of 14 in 8qxu
Go back to
Potassium binding site 7 out
of 14 in the In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
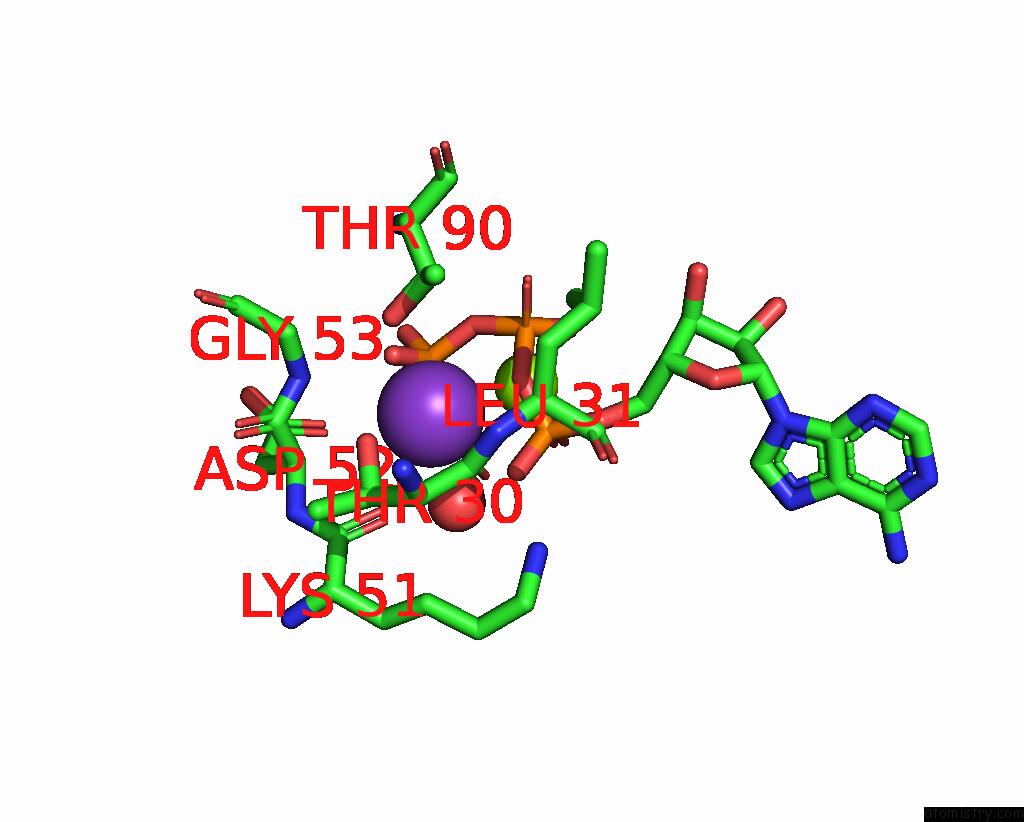
Mono view
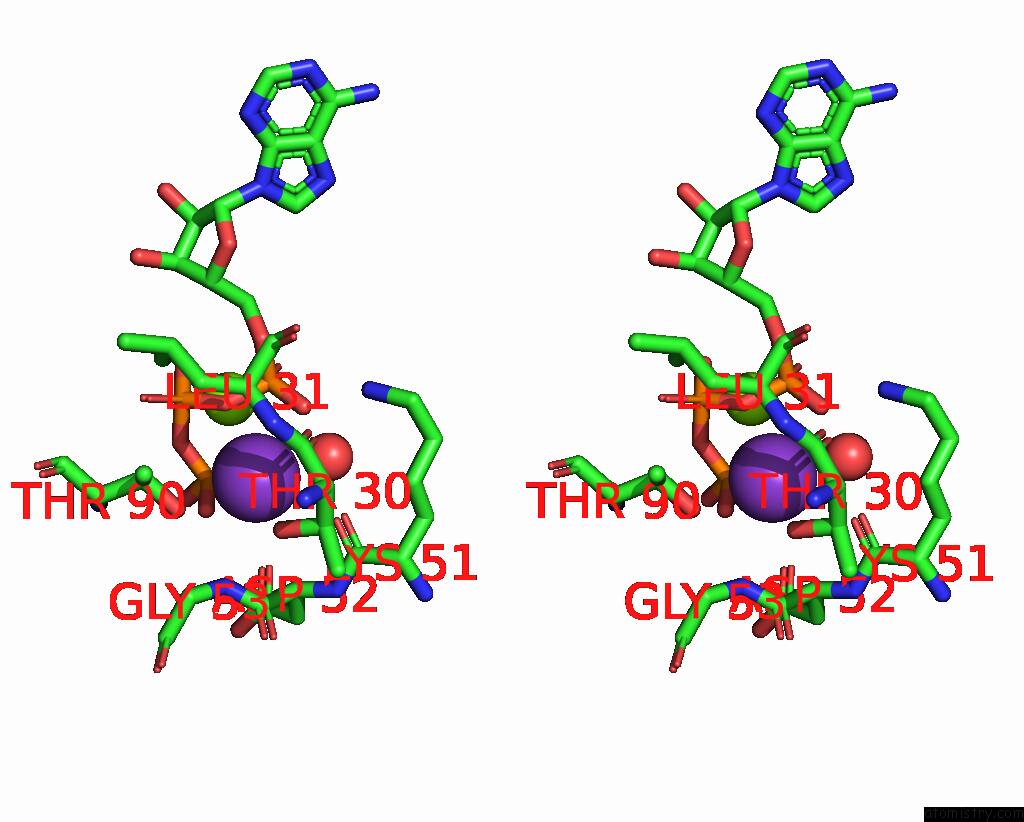
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 7 of In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography within 5.0Å range:
|
Potassium binding site 8 out of 14 in 8qxu
Go back to
Potassium binding site 8 out
of 14 in the In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
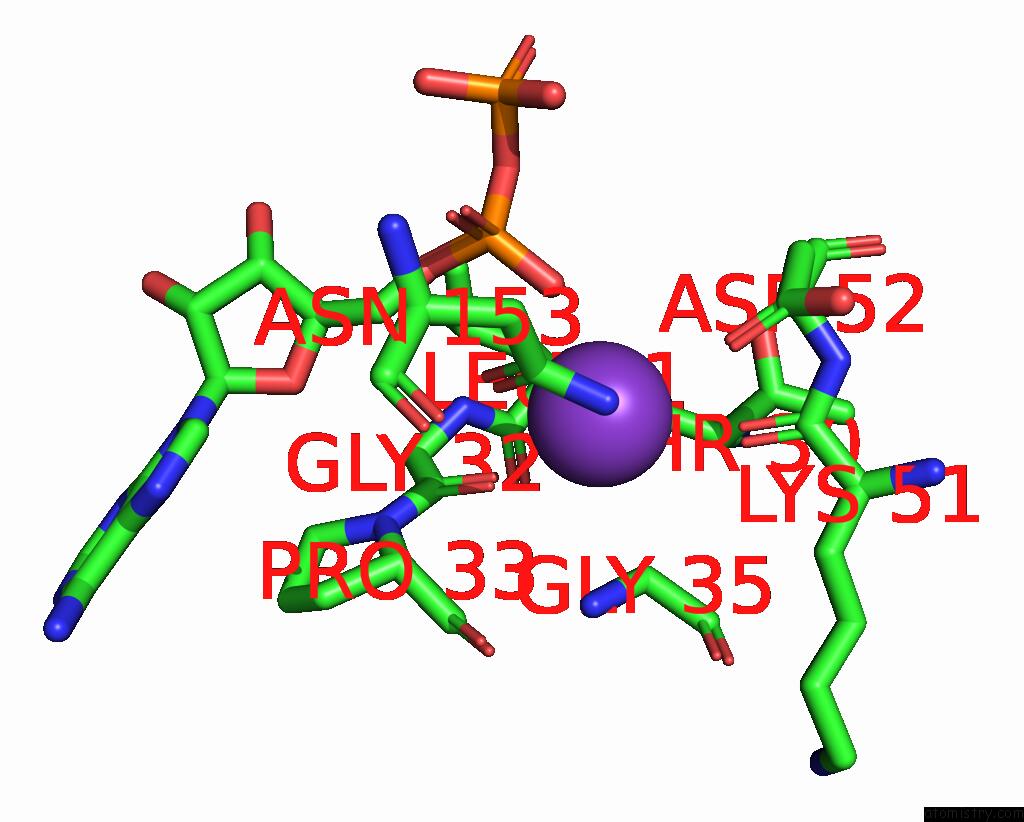
Mono view
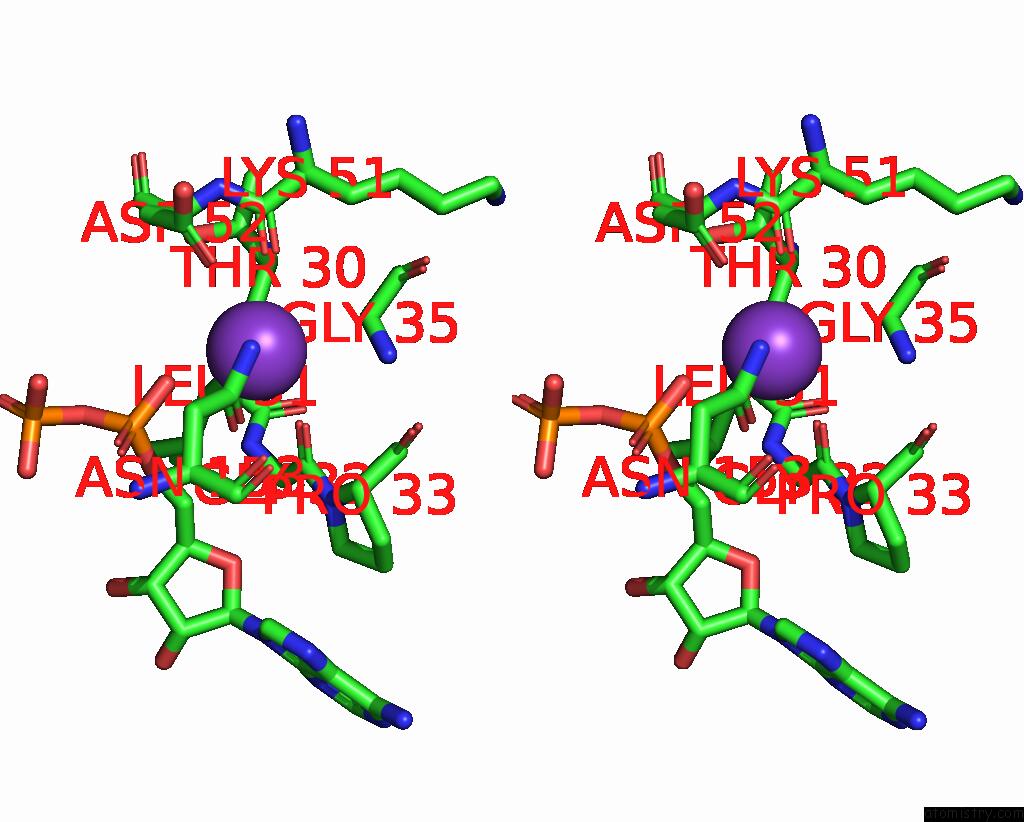
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 8 of In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography within 5.0Å range:
|
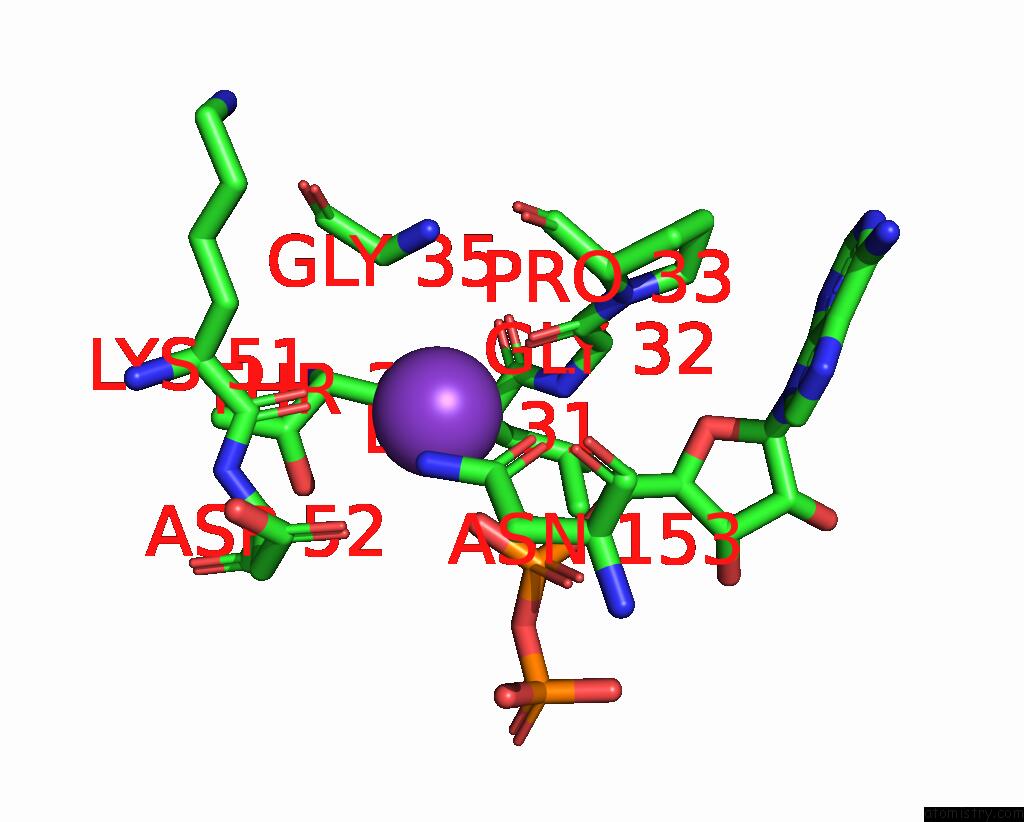
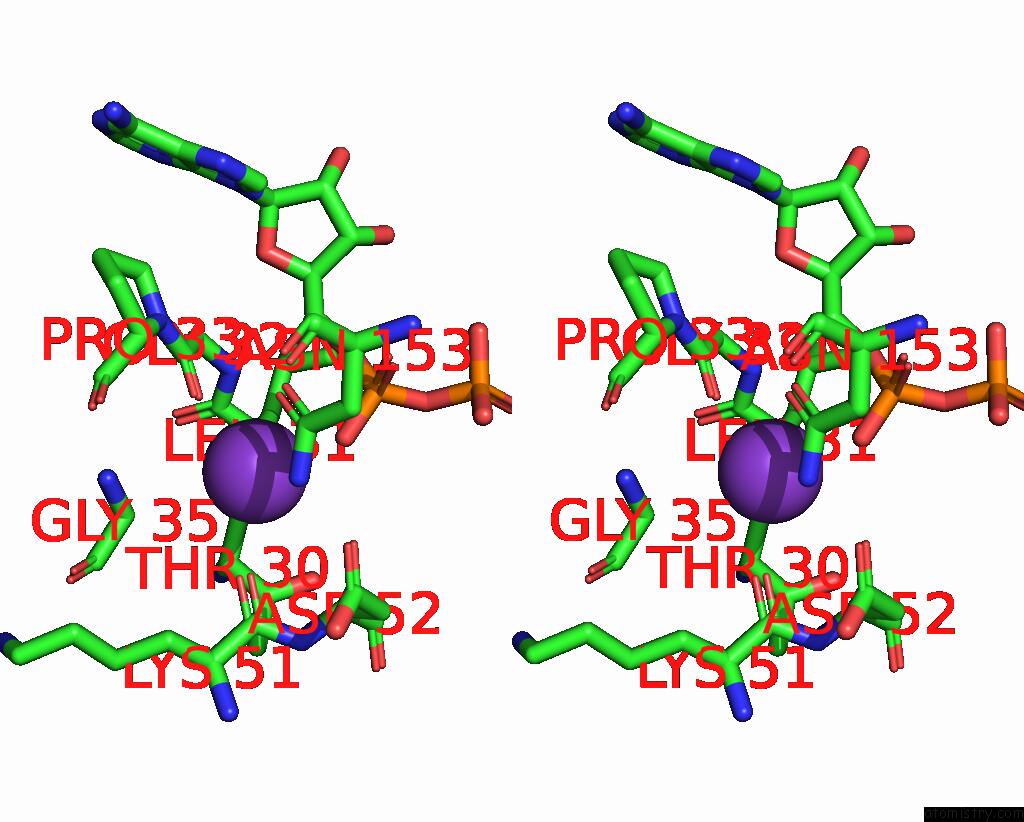
Potassium binding site 9 out of 14 in 8qxu
Go back to
Potassium binding site 9 out
of 14 in the In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 9 of In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography within 5.0Å range:
|
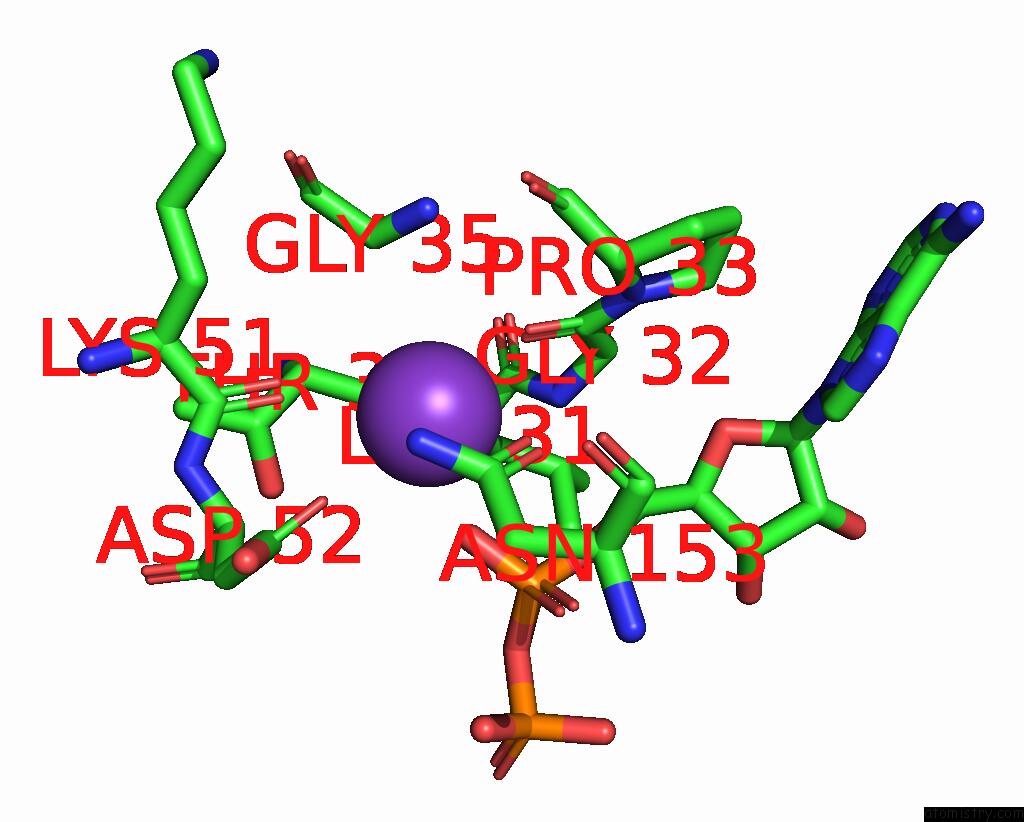
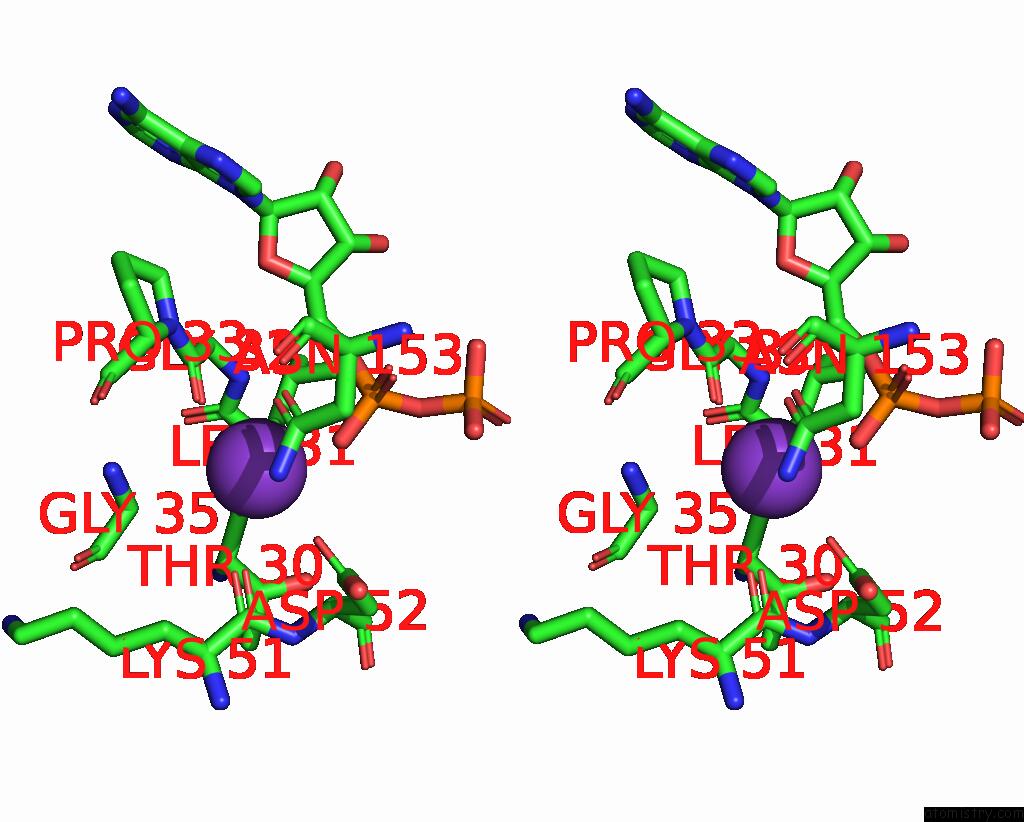
Potassium binding site 10 out of 14 in 8qxu
Go back to
Potassium binding site 10 out
of 14 in the In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 10 of In Situ Structure Average of GROEL14-GROES7 Complexes with Wide GROEL7 Trans Ring Conformation in Escherichia Coli Cytosol Obtained By Cryo Electron Tomography within 5.0Å range:
|
Reference:
J.Wagner,
A.I.Caravajal,
F.Beck,
A.Bracher,
W.Wan,
S.Bohn,
R.Koerner,
W.Baumeister,
R.Fernandez-Busnadiego,
F.U.Hartl.
Visualizing Chaperonin Function in Situ By Cryo-Electron Tomography Nature 2024.
ISSN: ESSN 1476-4687
Page generated: Sat Aug 9 17:35:25 2025
ISSN: ESSN 1476-4687
Last articles
Mg in 1VQPMg in 1VQO
Mg in 1W55
Mg in 1W54
Mg in 1W4B
Mg in 1W49
Mg in 1W46
Mg in 1W2Y
Mg in 1VQN
Mg in 1W25