Potassium »
PDB 8ooi-8qos »
8q1y »
Potassium in PDB 8q1y: Outward-Facing, OPEN2 Proteoliposome Complex I at 2.6 A, After Deactivation Treatment. Initially Purified in Lmng.
Enzymatic activity of Outward-Facing, OPEN2 Proteoliposome Complex I at 2.6 A, After Deactivation Treatment. Initially Purified in Lmng.
All present enzymatic activity of Outward-Facing, OPEN2 Proteoliposome Complex I at 2.6 A, After Deactivation Treatment. Initially Purified in Lmng.:
1.6.5.3; 1.6.99.3; 7.1.1.2;
1.6.5.3; 1.6.99.3; 7.1.1.2;
Other elements in 8q1y:
The structure of Outward-Facing, OPEN2 Proteoliposome Complex I at 2.6 A, After Deactivation Treatment. Initially Purified in Lmng. also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
| Iron | (Fe) | 28 atoms |
| Zinc | (Zn) | 2 atoms |
Potassium Binding Sites:
The binding sites of Potassium atom in the Outward-Facing, OPEN2 Proteoliposome Complex I at 2.6 A, After Deactivation Treatment. Initially Purified in Lmng.
(pdb code 8q1y). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total only one binding site of Potassium was determined in the Outward-Facing, OPEN2 Proteoliposome Complex I at 2.6 A, After Deactivation Treatment. Initially Purified in Lmng., PDB code: 8q1y:
In total only one binding site of Potassium was determined in the Outward-Facing, OPEN2 Proteoliposome Complex I at 2.6 A, After Deactivation Treatment. Initially Purified in Lmng., PDB code: 8q1y:
Potassium binding site 1 out of 1 in 8q1y
Go back to
Potassium binding site 1 out
of 1 in the Outward-Facing, OPEN2 Proteoliposome Complex I at 2.6 A, After Deactivation Treatment. Initially Purified in Lmng.
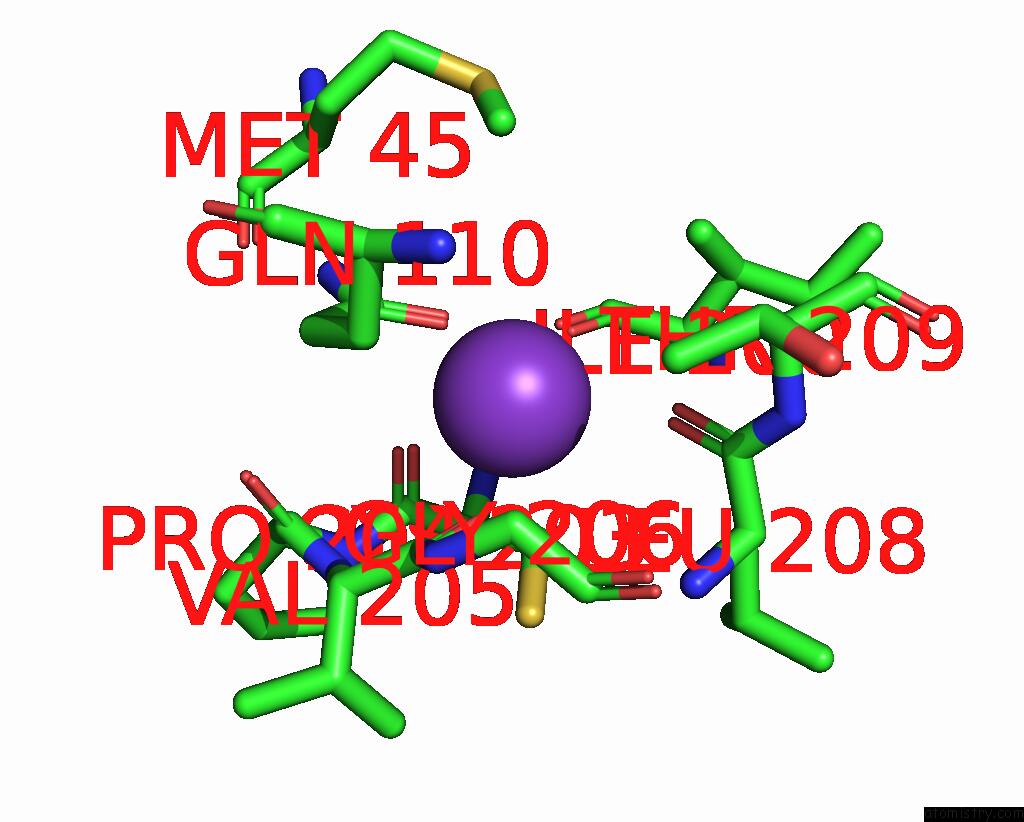
Mono view
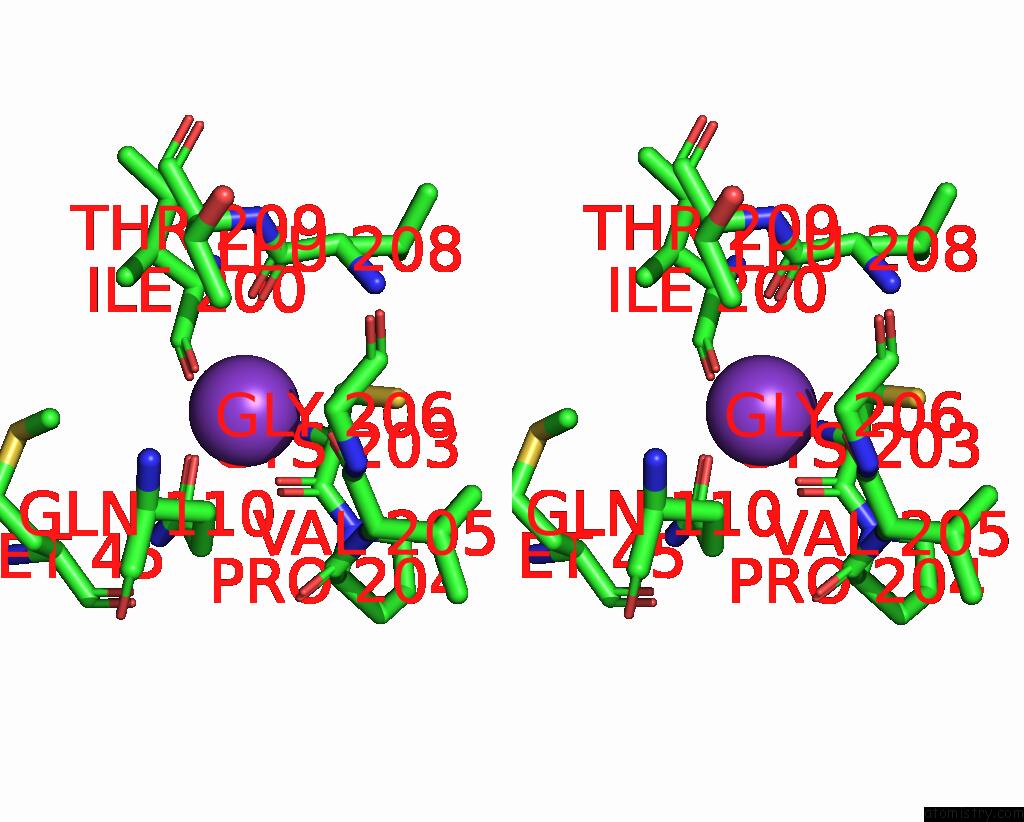
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Outward-Facing, OPEN2 Proteoliposome Complex I at 2.6 A, After Deactivation Treatment. Initially Purified in Lmng. within 5.0Å range:
|
Reference:
D.N.Grba,
J.J.Wright,
W.Fisher,
Z.Yin,
J.Hirst.
Molecular Mechanism of the Ischemia-Induced Regulatory Switch in Mammalian Complex I Science 2024.
ISSN: ESSN 1095-9203
DOI: 10.1126/SCIENCE.ADO2075
Page generated: Sat Aug 9 17:27:51 2025
ISSN: ESSN 1095-9203
DOI: 10.1126/SCIENCE.ADO2075
Last articles
Mg in 1XNGMg in 1XN0
Mg in 1XMY
Mg in 1XMV
Mg in 1XMX
Mg in 1XMI
Mg in 1XMU
Mg in 1XM6
Mg in 1XMJ
Mg in 1XM4