Potassium »
PDB 8dod-8fo0 »
8f6m »
Potassium in PDB 8f6m: Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex.
Enzymatic activity of Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex.
All present enzymatic activity of Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex.:
2.7.1.40; 2.7.10.2; 2.7.11.1;
2.7.1.40; 2.7.10.2; 2.7.11.1;
Protein crystallography data
The structure of Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex., PDB code: 8f6m
was solved by
T.Holyoak,
A.W.Fenton,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.02 / 2.15 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 93.026, 216.396, 258.151, 90, 90, 90 |
| R / Rfree (%) | 15.6 / 20.5 |
Other elements in 8f6m:
The structure of Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex. also contains other interesting chemical elements:
| Manganese | (Mn) | 15 atoms |
Potassium Binding Sites:
The binding sites of Potassium atom in the Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex.
(pdb code 8f6m). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 8 binding sites of Potassium where determined in the Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex., PDB code: 8f6m:
Jump to Potassium binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Potassium where determined in the Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex., PDB code: 8f6m:
Jump to Potassium binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Potassium binding site 1 out of 8 in 8f6m
Go back to
Potassium binding site 1 out
of 8 in the Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex.
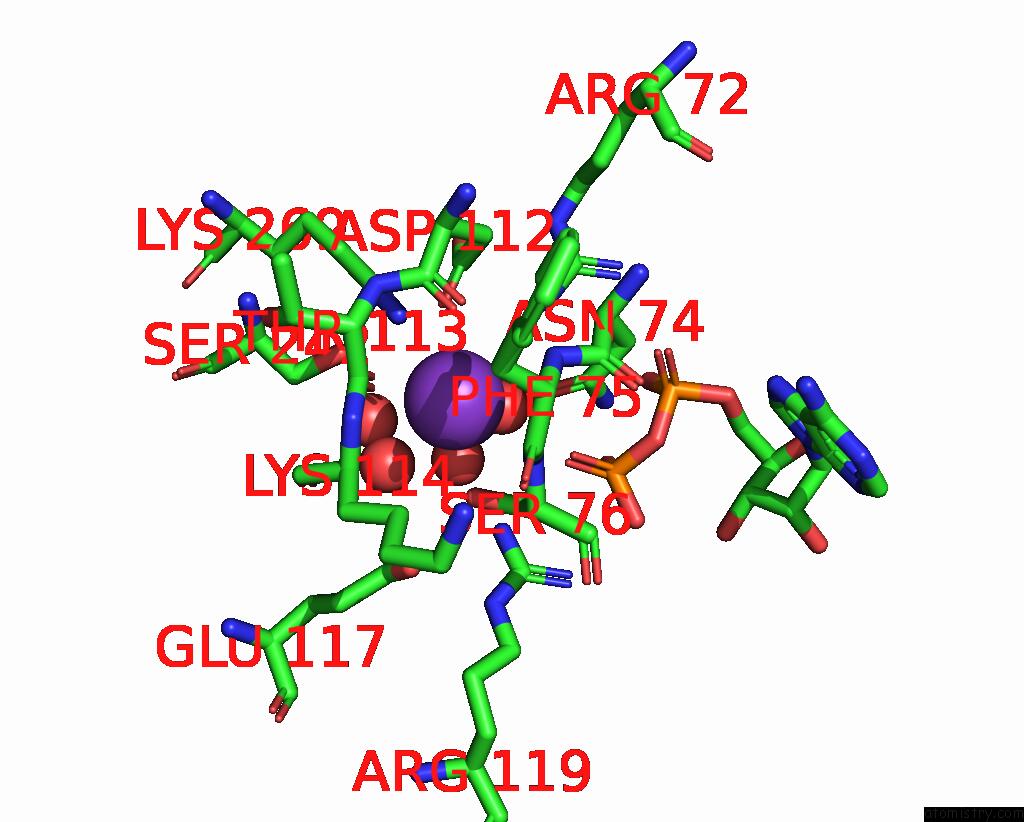
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex. within 5.0Å range:
|
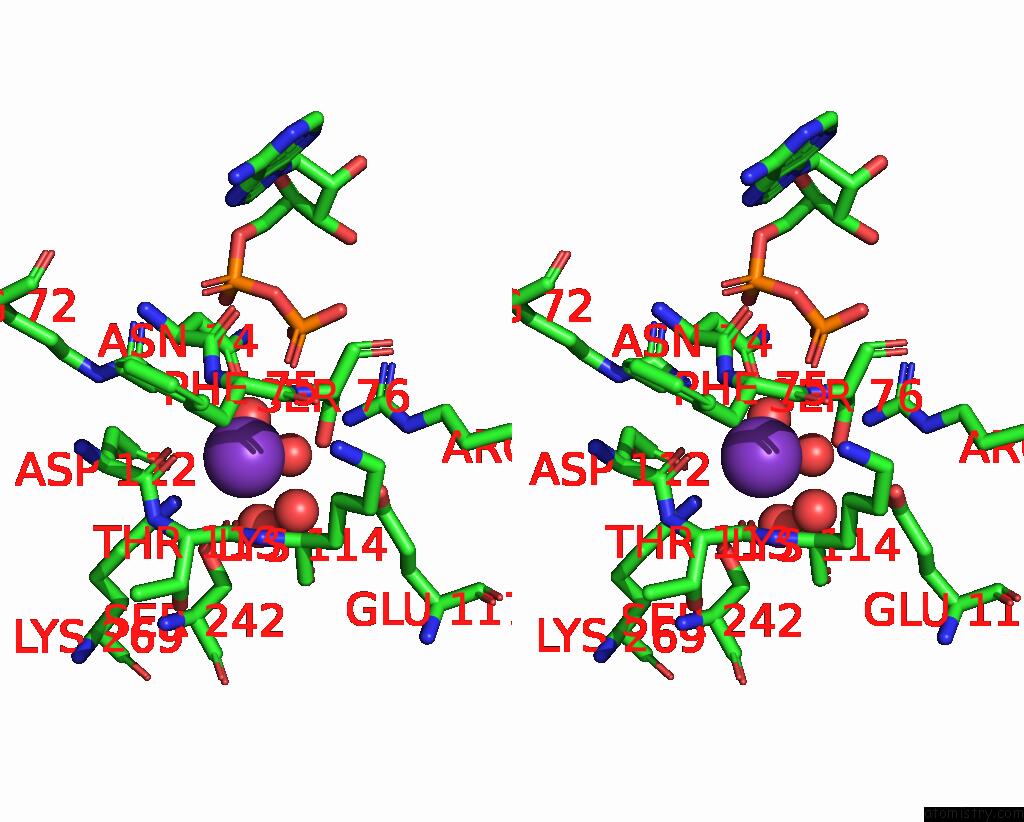
Potassium binding site 2 out of 8 in 8f6m
Go back to
Potassium binding site 2 out
of 8 in the Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex.
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex. within 5.0Å range:
|
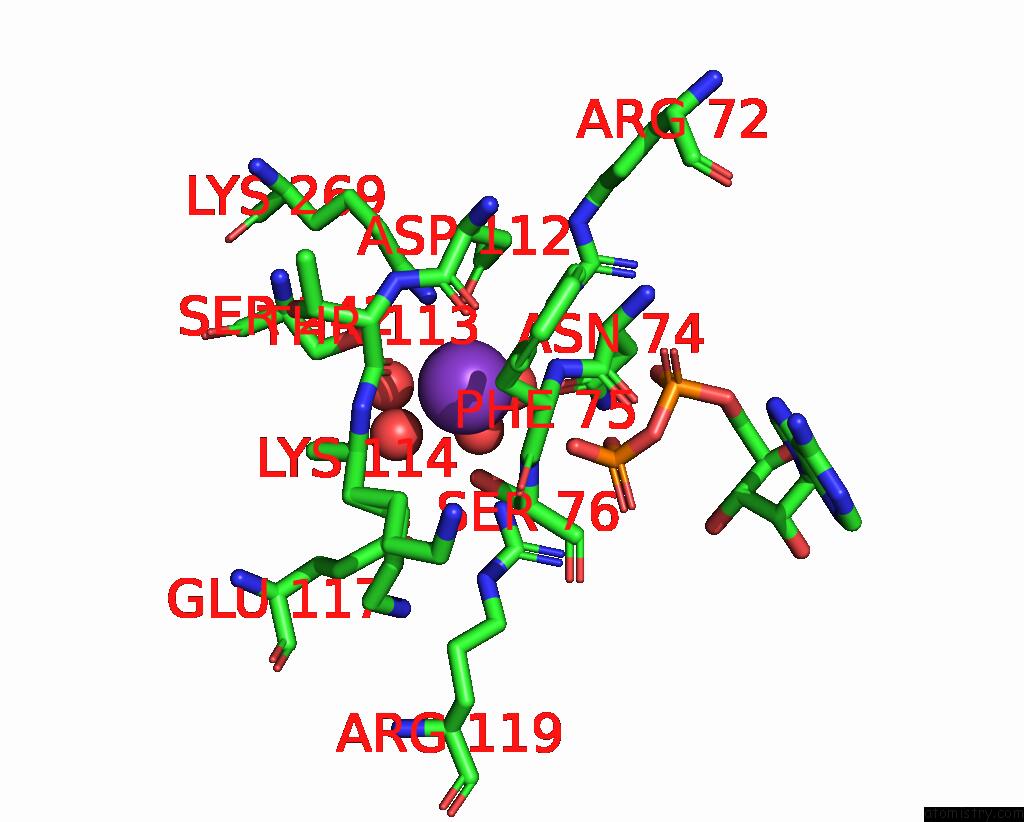
Potassium binding site 3 out of 8 in 8f6m
Go back to
Potassium binding site 3 out
of 8 in the Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex.
Mono view
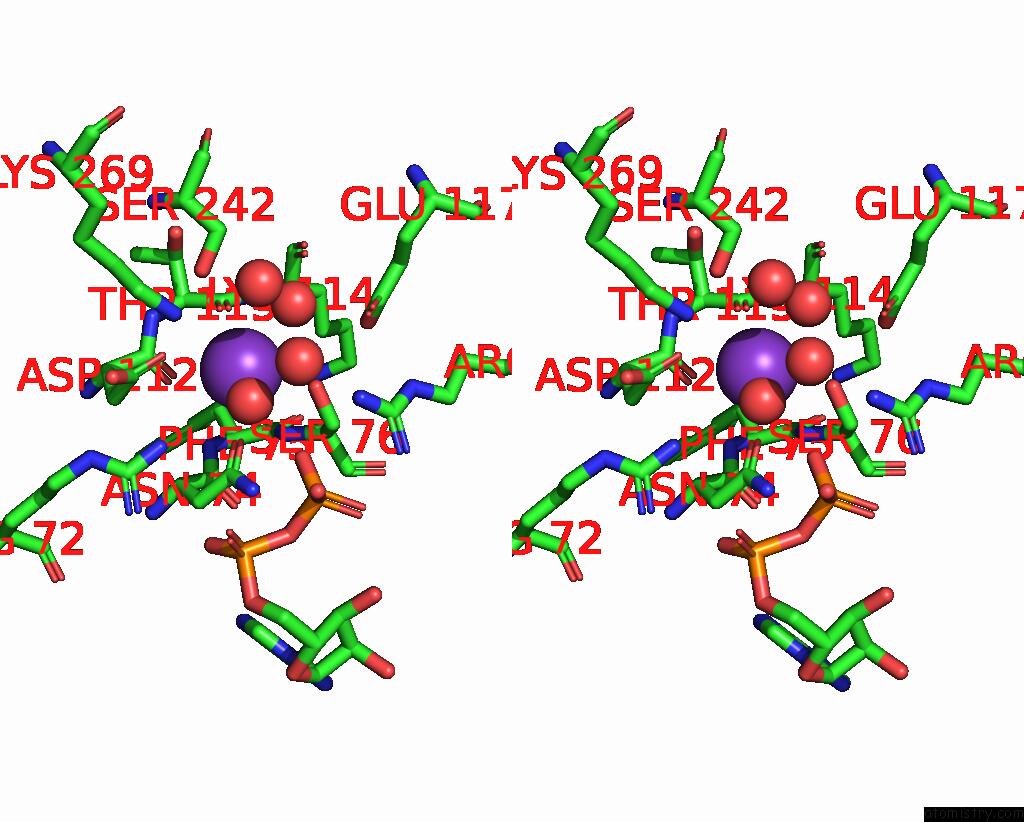
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 3 of Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex. within 5.0Å range:
|
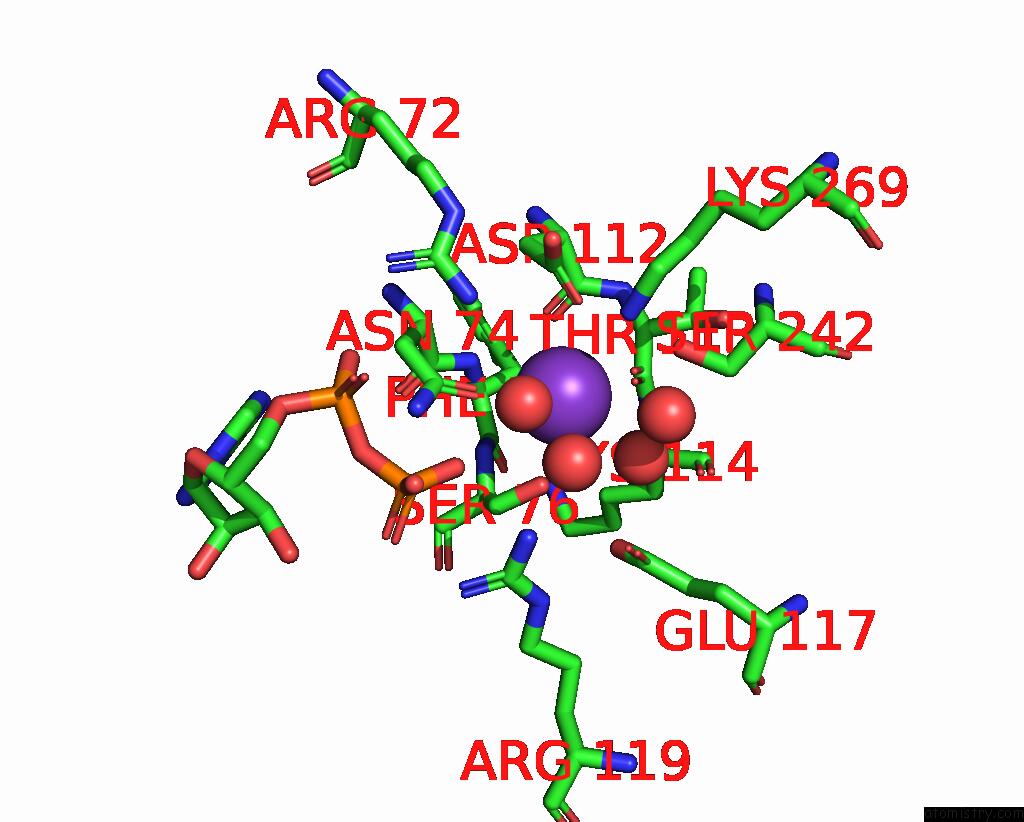
Potassium binding site 4 out of 8 in 8f6m
Go back to
Potassium binding site 4 out
of 8 in the Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex.
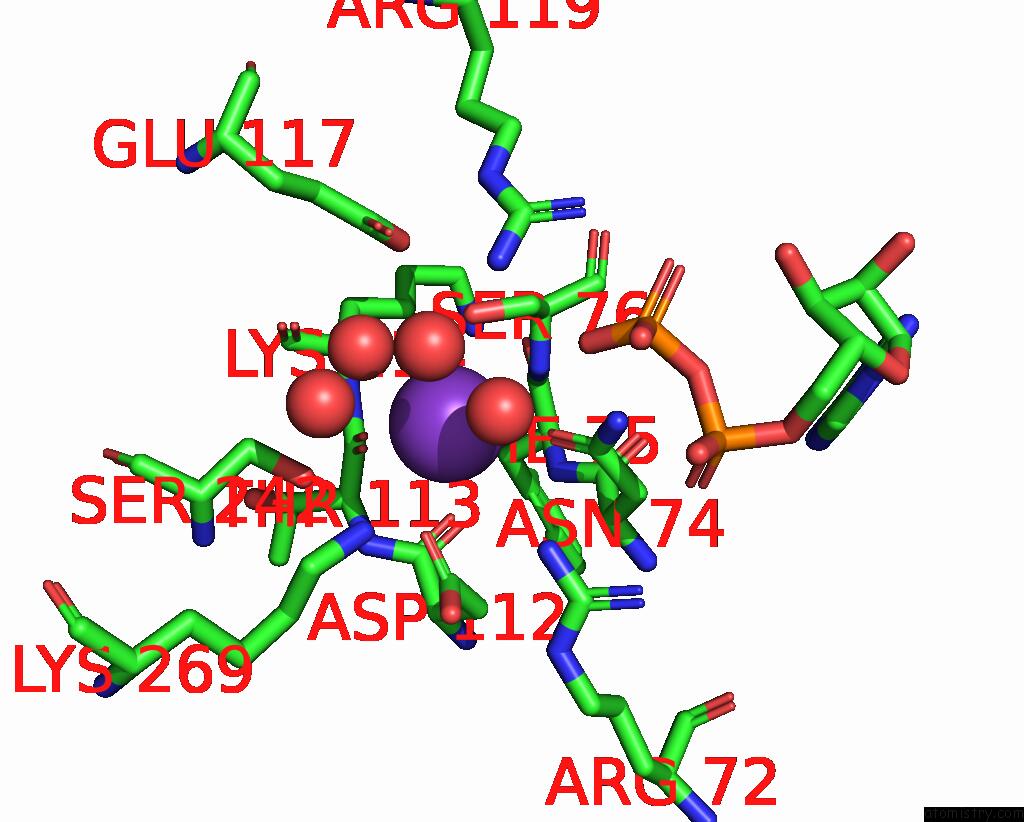
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 4 of Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex. within 5.0Å range:
|
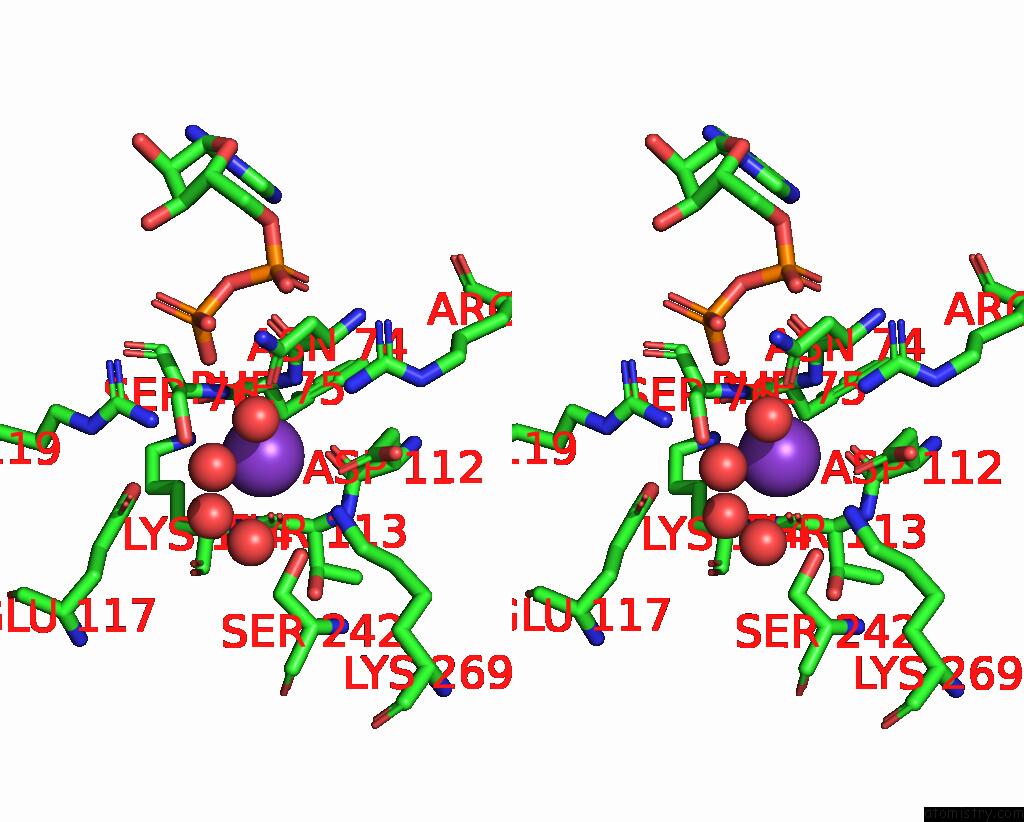
Potassium binding site 5 out of 8 in 8f6m
Go back to
Potassium binding site 5 out
of 8 in the Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex.
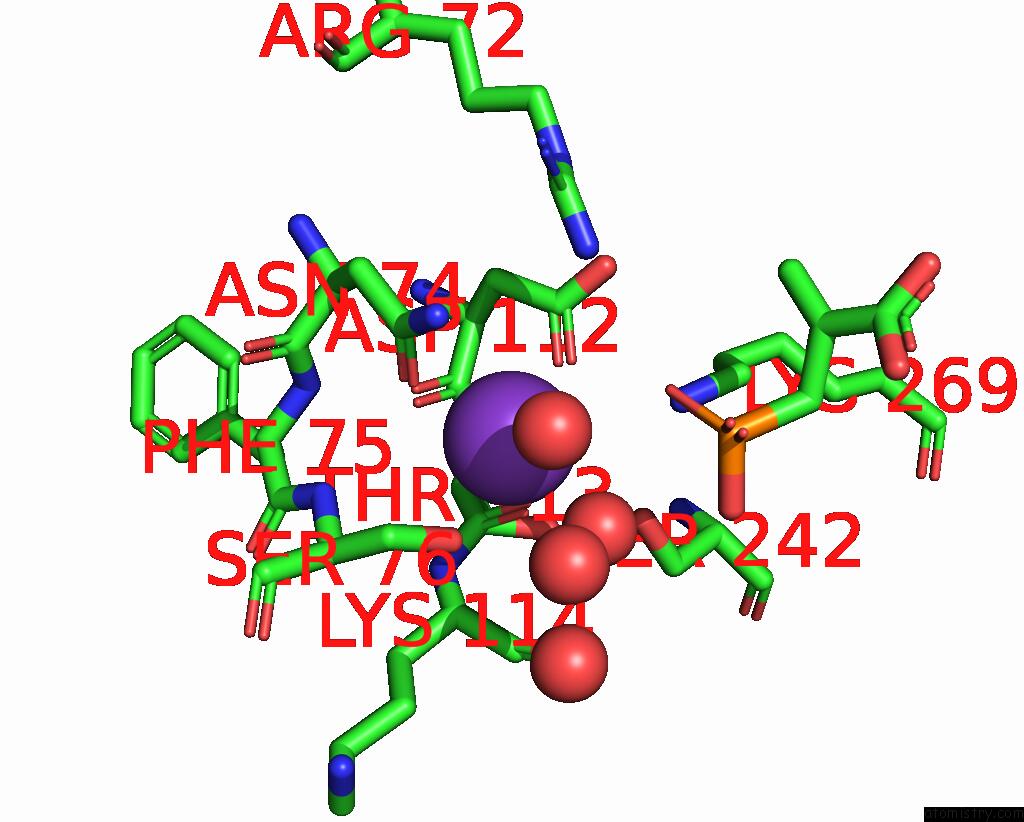
Mono view
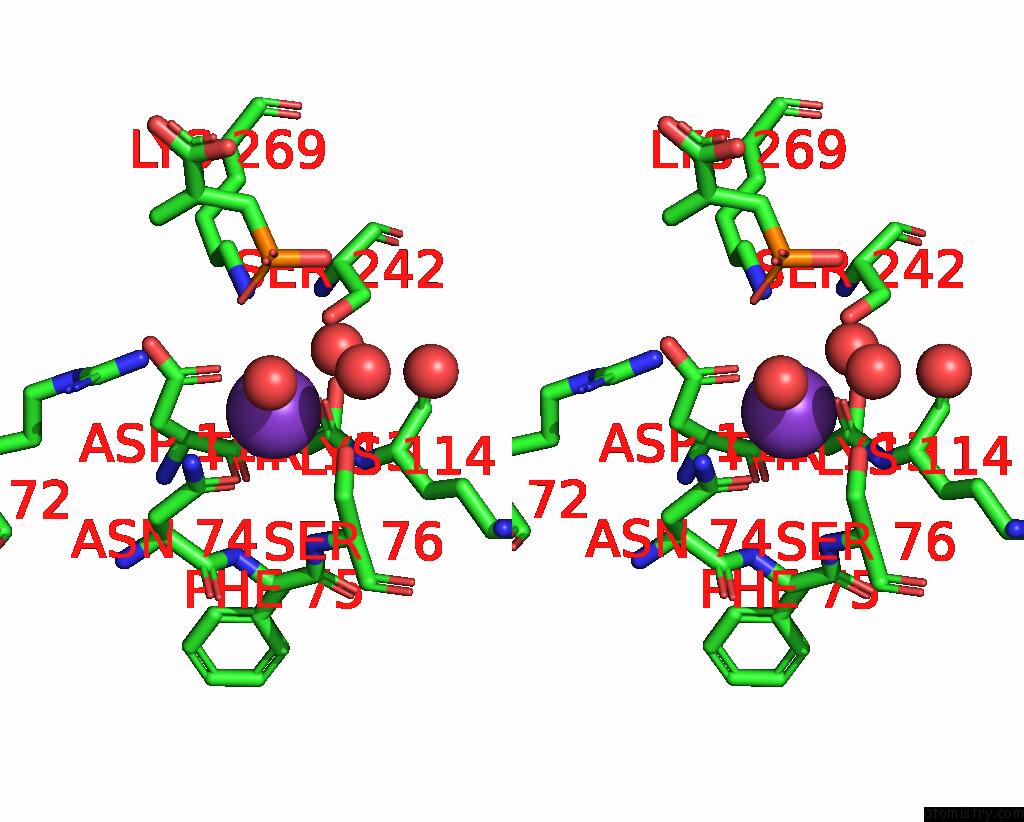
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 5 of Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex. within 5.0Å range:
|
Potassium binding site 6 out of 8 in 8f6m
Go back to
Potassium binding site 6 out
of 8 in the Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex.
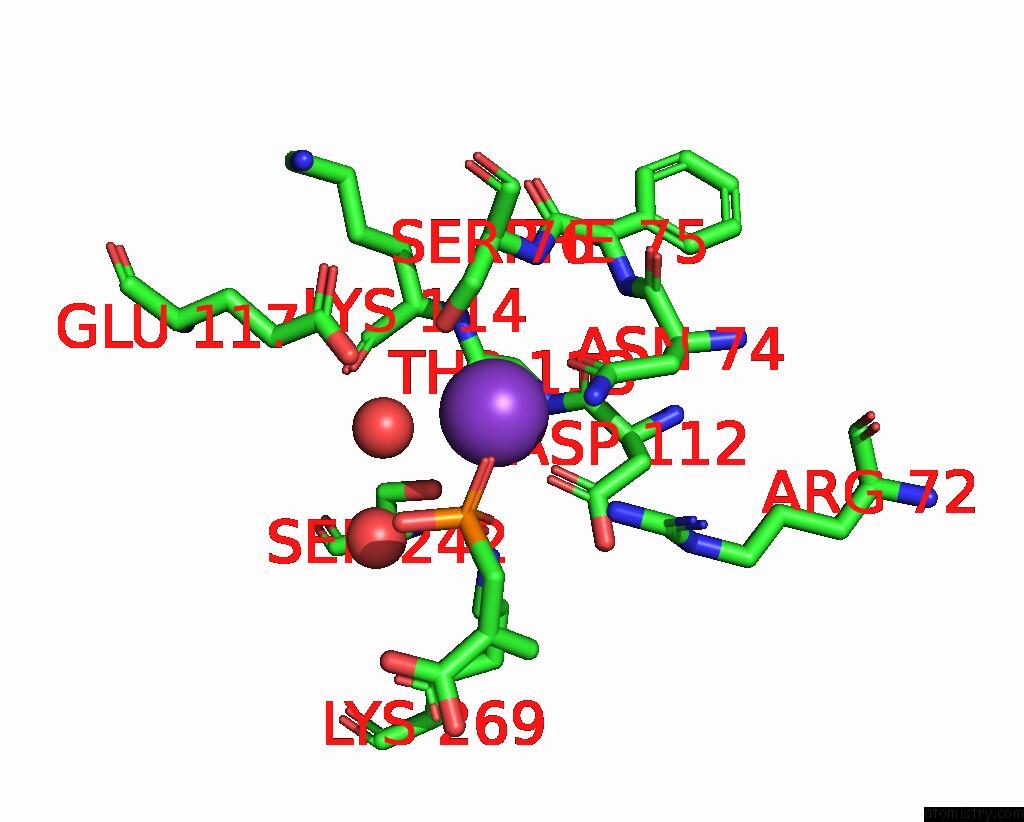
Mono view
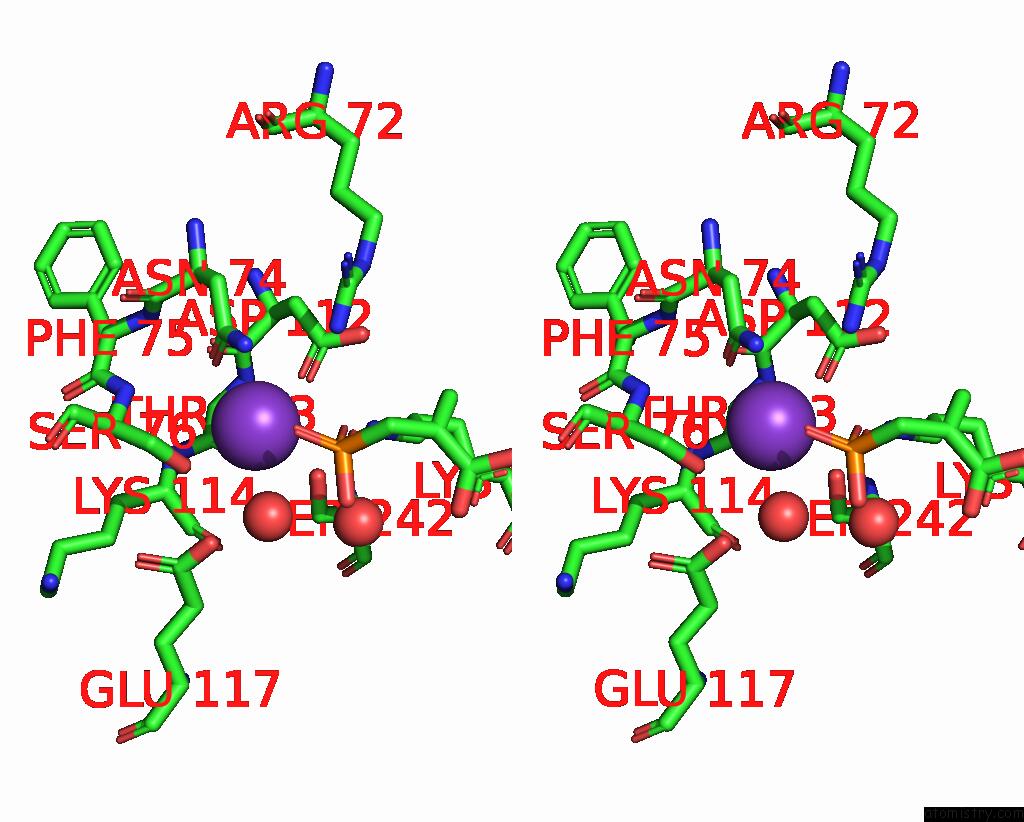
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 6 of Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex. within 5.0Å range:
|
Potassium binding site 7 out of 8 in 8f6m
Go back to
Potassium binding site 7 out
of 8 in the Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex.

Mono view
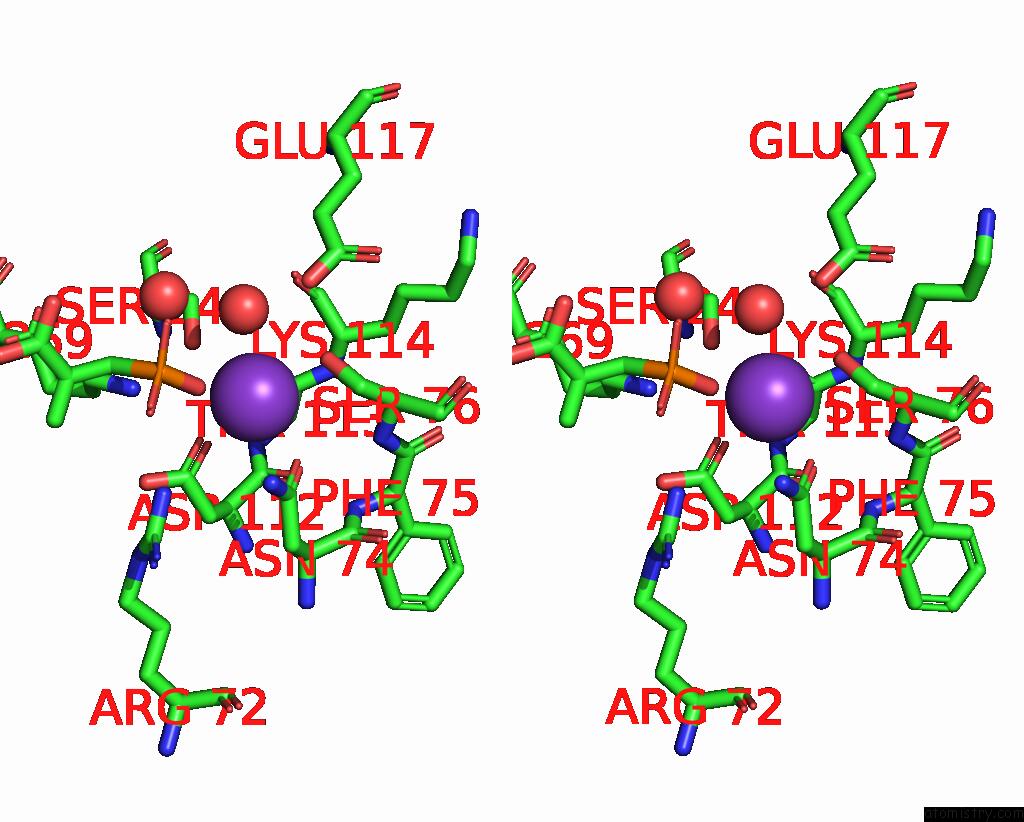
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 7 of Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex. within 5.0Å range:
|
Potassium binding site 8 out of 8 in 8f6m
Go back to
Potassium binding site 8 out
of 8 in the Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex.

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 8 of Complex of Rabbit Muscle Pyruvate Kinase with Adp and the Phosphonate Analogue of Pep Mimicking the Michaelis Complex. within 5.0Å range:
|
Reference:
L.Swint-Kruse,
L.L.Dougherty,
B.Page,
T.Wu,
P.T.O'neil,
C.B.Prasannan,
C.Timmons,
Q.Tang,
D.J.Parente,
S.Sreenivasan,
T.Holyoak,
A.W.Fenton.
Pyk-Substitutionome: An Integrated Database Containing Allosteric Coupling, Ligand Affinity and Mutational, Structural, Pathological, Bioinformatic and Computational Information About Pyruvate Kinase Isozymes. Database (Oxford) V.2023 2023.
ISSN: ISSN 1758-0463
PubMed: 37171062
DOI: 10.1093/DATABASE/BAAD030
Page generated: Sat Aug 9 16:50:32 2025
ISSN: ISSN 1758-0463
PubMed: 37171062
DOI: 10.1093/DATABASE/BAAD030
Last articles
Mg in 2VX1Mg in 2VX0
Mg in 2VWZ
Mg in 2VXT
Mg in 2VWX
Mg in 2VWY
Mg in 2VWT
Mg in 2VWI
Mg in 2VVG
Mg in 2VUI