Potassium »
PDB 8cxz-8eh1 »
8de7 »
Potassium in PDB 8de7: Cryo-Em Structure of the Zebrafish Two Pore Domain K+ Channel TREK1 (K2P2.1) in Ddm Detergent
Potassium Binding Sites:
The binding sites of Potassium atom in the Cryo-Em Structure of the Zebrafish Two Pore Domain K+ Channel TREK1 (K2P2.1) in Ddm Detergent
(pdb code 8de7). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 3 binding sites of Potassium where determined in the Cryo-Em Structure of the Zebrafish Two Pore Domain K+ Channel TREK1 (K2P2.1) in Ddm Detergent, PDB code: 8de7:
Jump to Potassium binding site number: 1; 2; 3;
In total 3 binding sites of Potassium where determined in the Cryo-Em Structure of the Zebrafish Two Pore Domain K+ Channel TREK1 (K2P2.1) in Ddm Detergent, PDB code: 8de7:
Jump to Potassium binding site number: 1; 2; 3;
Potassium binding site 1 out of 3 in 8de7
Go back to
Potassium binding site 1 out
of 3 in the Cryo-Em Structure of the Zebrafish Two Pore Domain K+ Channel TREK1 (K2P2.1) in Ddm Detergent
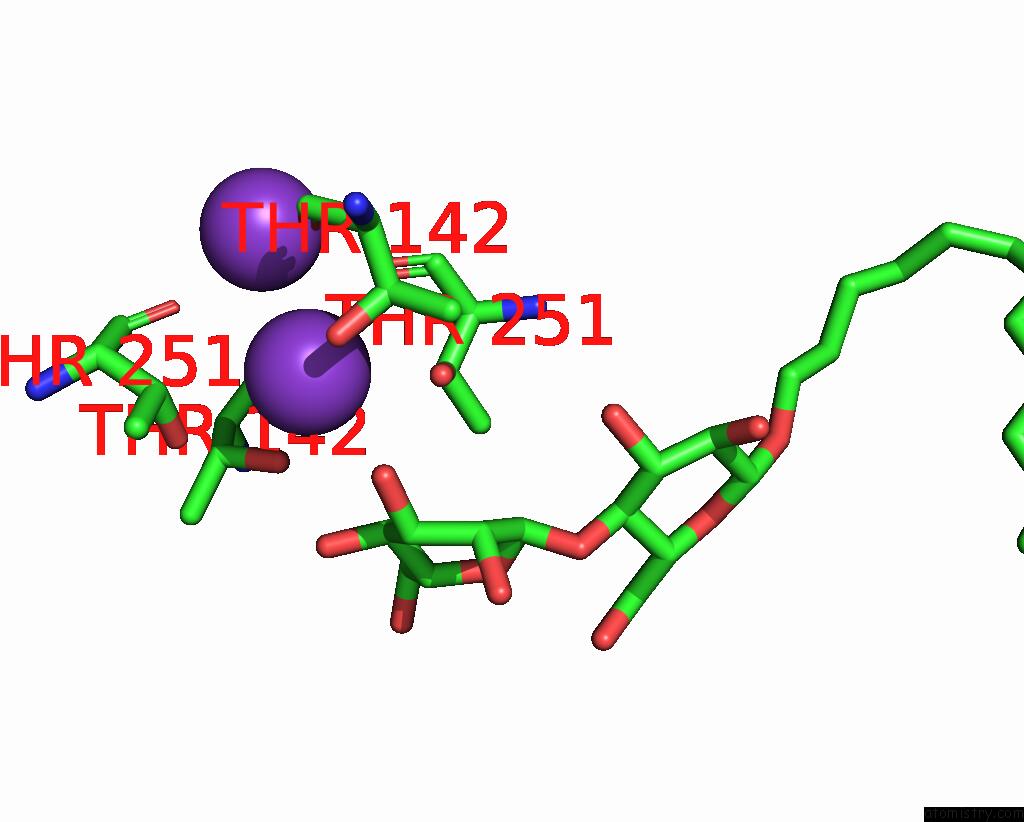
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Cryo-Em Structure of the Zebrafish Two Pore Domain K+ Channel TREK1 (K2P2.1) in Ddm Detergent within 5.0Å range:
|
Potassium binding site 2 out of 3 in 8de7
Go back to
Potassium binding site 2 out
of 3 in the Cryo-Em Structure of the Zebrafish Two Pore Domain K+ Channel TREK1 (K2P2.1) in Ddm Detergent
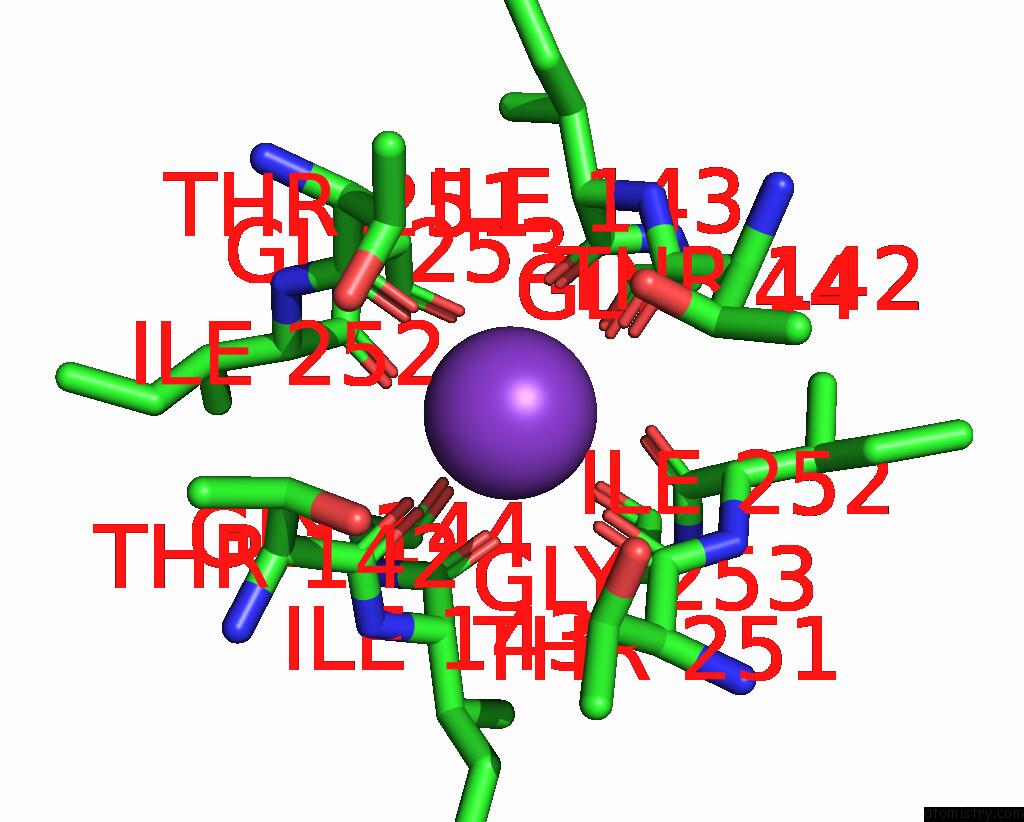
Mono view
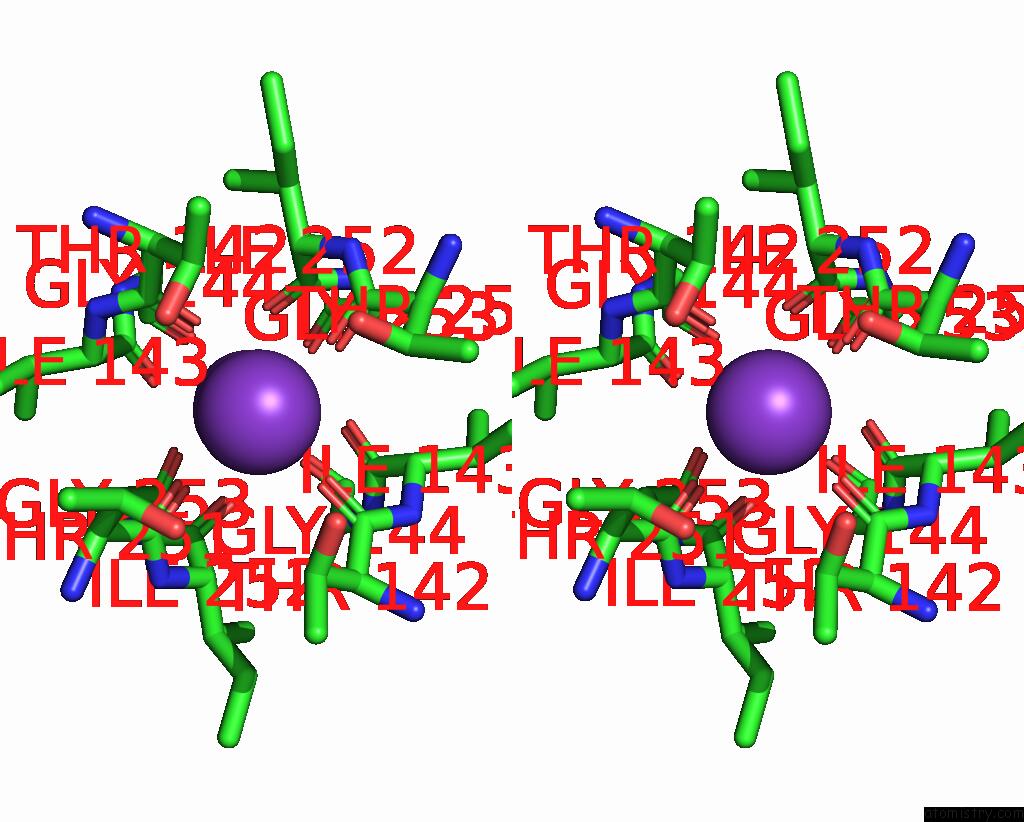
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Cryo-Em Structure of the Zebrafish Two Pore Domain K+ Channel TREK1 (K2P2.1) in Ddm Detergent within 5.0Å range:
|
Potassium binding site 3 out of 3 in 8de7
Go back to
Potassium binding site 3 out
of 3 in the Cryo-Em Structure of the Zebrafish Two Pore Domain K+ Channel TREK1 (K2P2.1) in Ddm Detergent
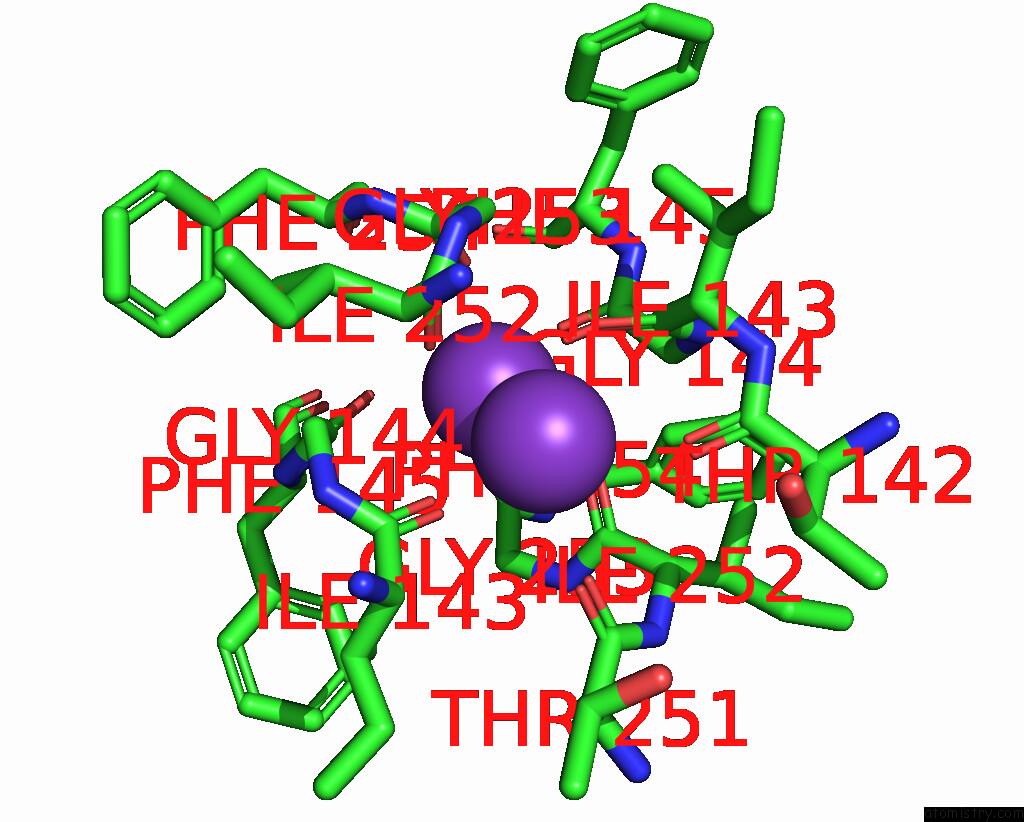
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 3 of Cryo-Em Structure of the Zebrafish Two Pore Domain K+ Channel TREK1 (K2P2.1) in Ddm Detergent within 5.0Å range:
|
Reference:
P.A.M.Schmidpeter,
J.T.Petroff 2Nd,
L.Khajoueinejad,
A.Wague,
C.Frankfater,
W.W.L.Cheng,
C.M.Nimigean,
P.M.Riegelhaupt.
Membrane Phospholipids Control Gating of the Mechanosensitive Potassium Leak Channel TREK1. Nat Commun V. 14 1077 2023.
ISSN: ESSN 2041-1723
PubMed: 36841877
DOI: 10.1038/S41467-023-36765-W
Page generated: Mon Aug 12 23:30:13 2024
ISSN: ESSN 2041-1723
PubMed: 36841877
DOI: 10.1038/S41467-023-36765-W
Last articles
Zn in 9JYWZn in 9IR4
Zn in 9IR3
Zn in 9GMX
Zn in 9GMW
Zn in 9JEJ
Zn in 9ERF
Zn in 9ERE
Zn in 9EGV
Zn in 9EGW