Potassium »
PDB 7zri-8bm0 »
7zri »
Potassium in PDB 7zri: Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+
Enzymatic activity of Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+
All present enzymatic activity of Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+:
7.2.2.6;
7.2.2.6;
Potassium Binding Sites:
The binding sites of Potassium atom in the Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+
(pdb code 7zri). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 7 binding sites of Potassium where determined in the Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+, PDB code: 7zri:
Jump to Potassium binding site number: 1; 2; 3; 4; 5; 6; 7;
In total 7 binding sites of Potassium where determined in the Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+, PDB code: 7zri:
Jump to Potassium binding site number: 1; 2; 3; 4; 5; 6; 7;
Potassium binding site 1 out of 7 in 7zri
Go back to
Potassium binding site 1 out
of 7 in the Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+ within 5.0Å range:
|
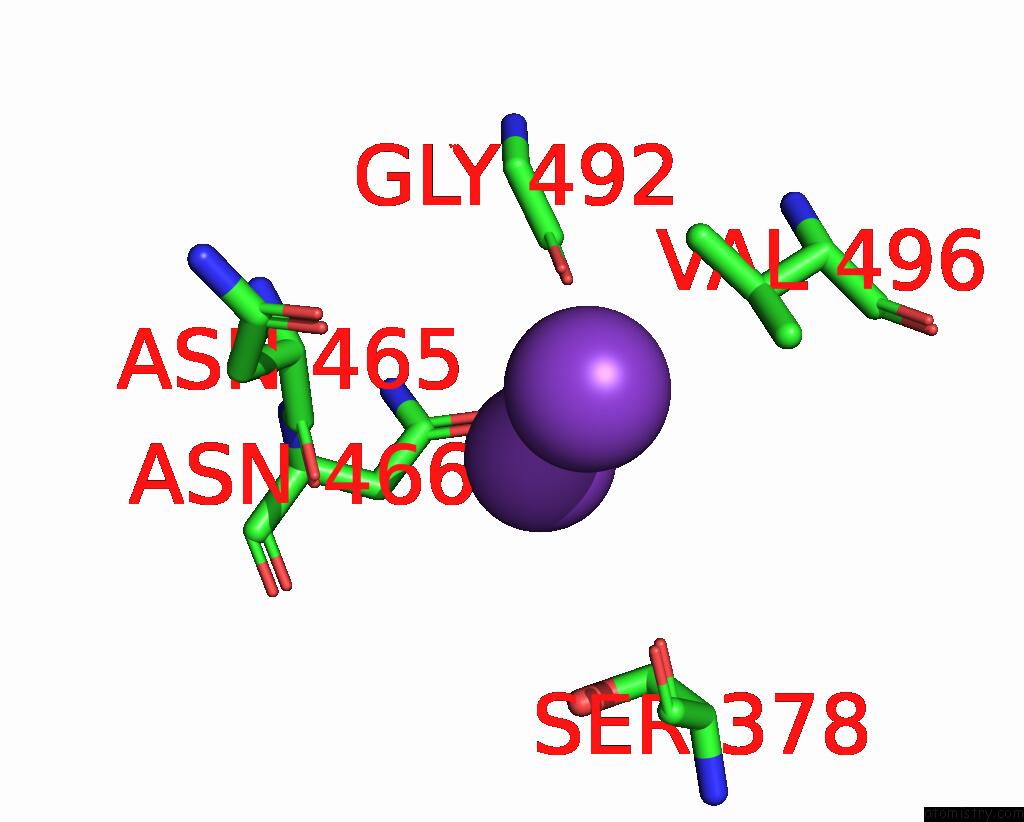
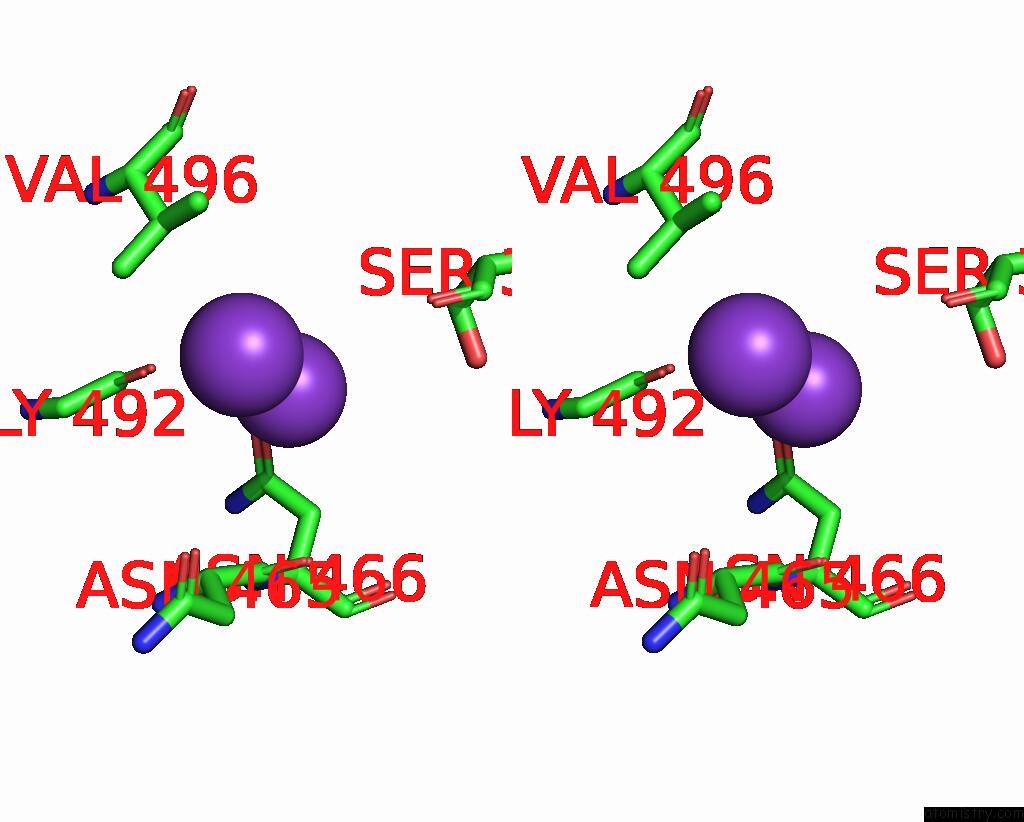
Potassium binding site 2 out of 7 in 7zri
Go back to
Potassium binding site 2 out
of 7 in the Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+ within 5.0Å range:
|
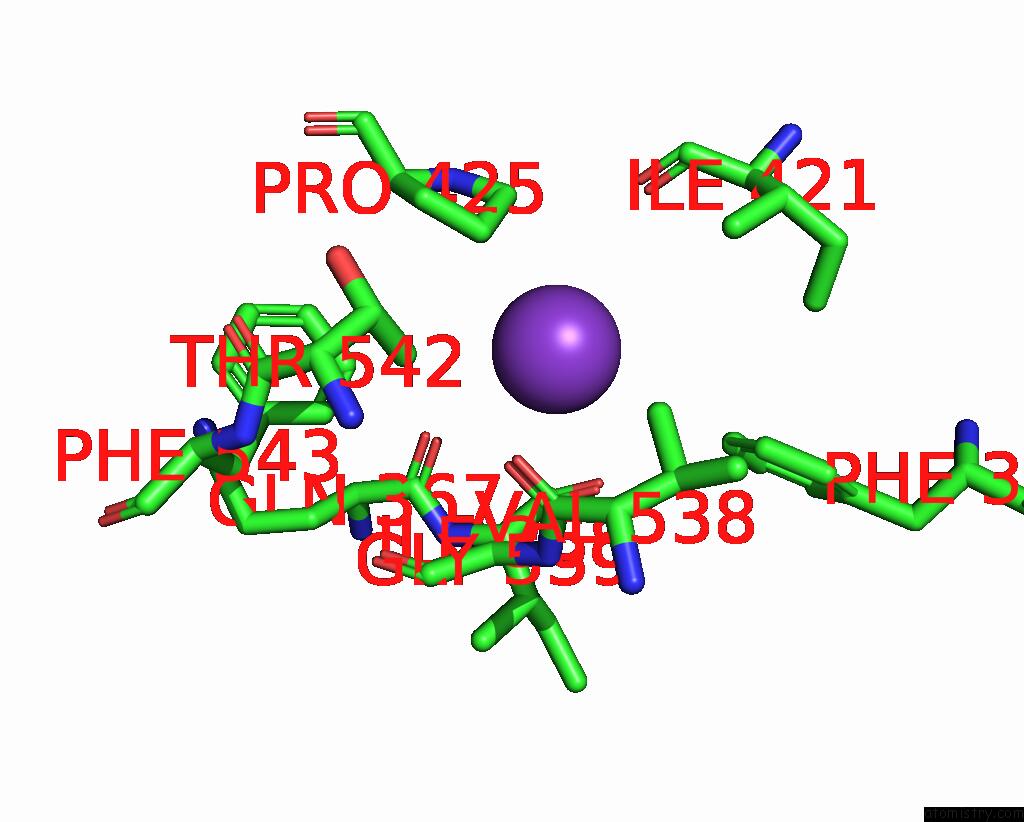
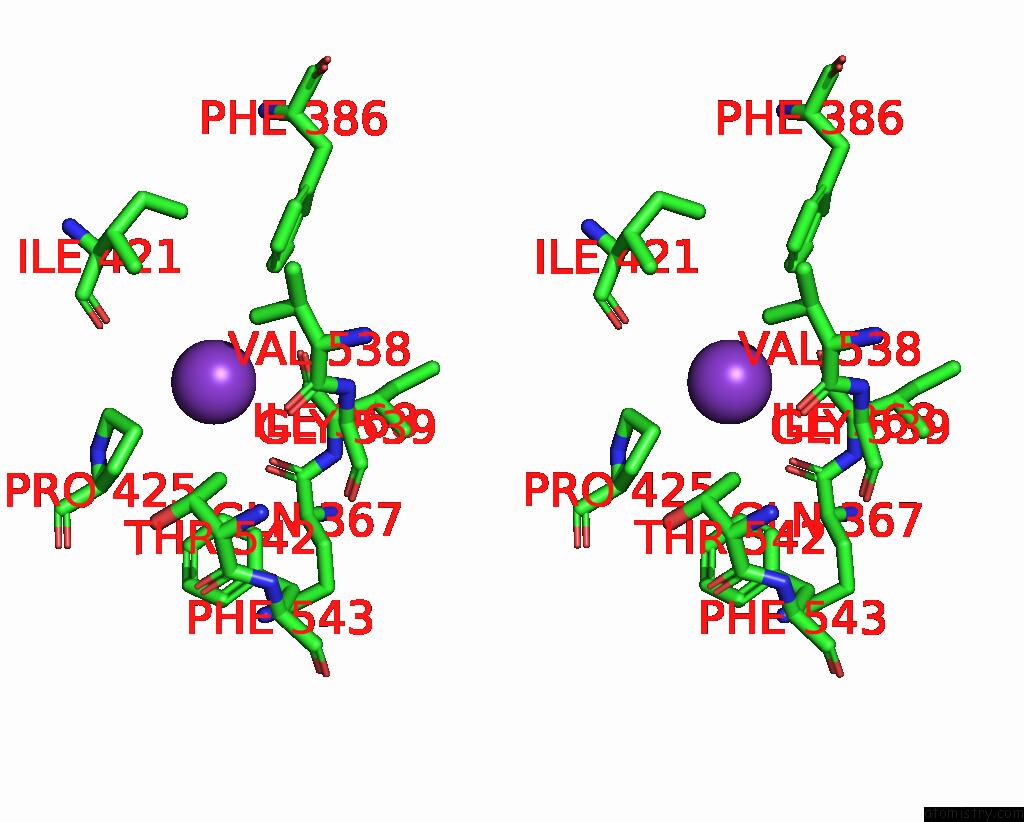
Potassium binding site 3 out of 7 in 7zri
Go back to
Potassium binding site 3 out
of 7 in the Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 3 of Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+ within 5.0Å range:
|
Potassium binding site 4 out of 7 in 7zri
Go back to
Potassium binding site 4 out
of 7 in the Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+
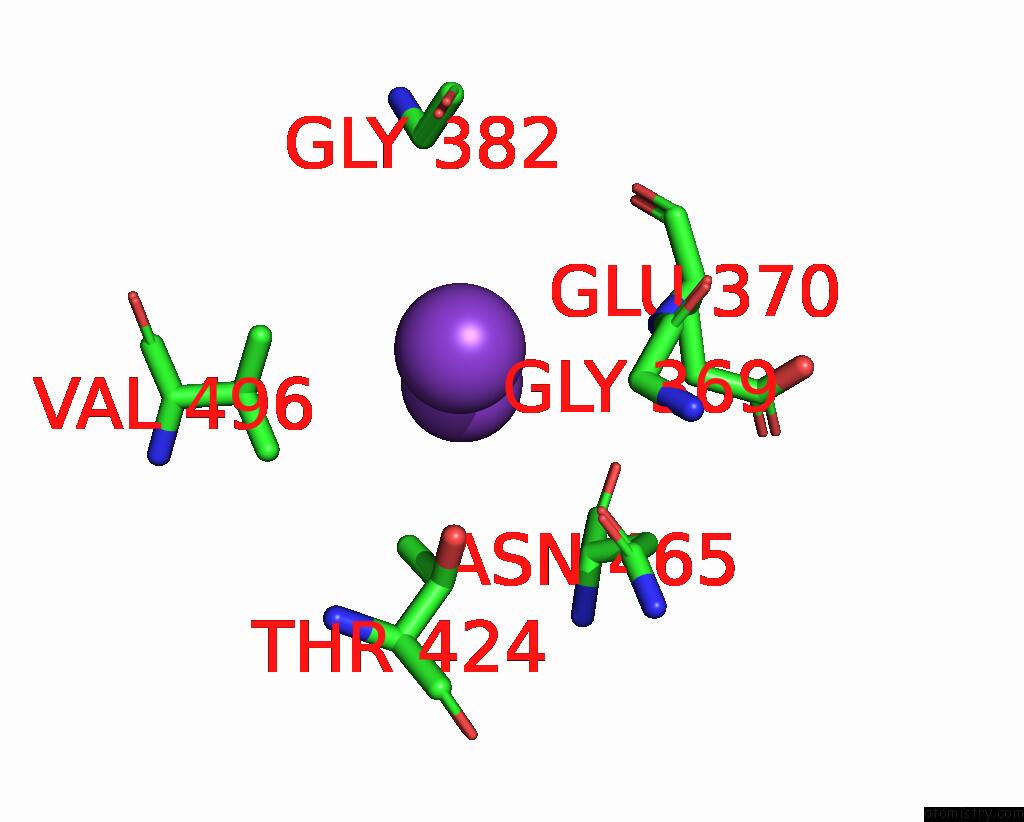
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 4 of Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+ within 5.0Å range:
|
Potassium binding site 5 out of 7 in 7zri
Go back to
Potassium binding site 5 out
of 7 in the Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+
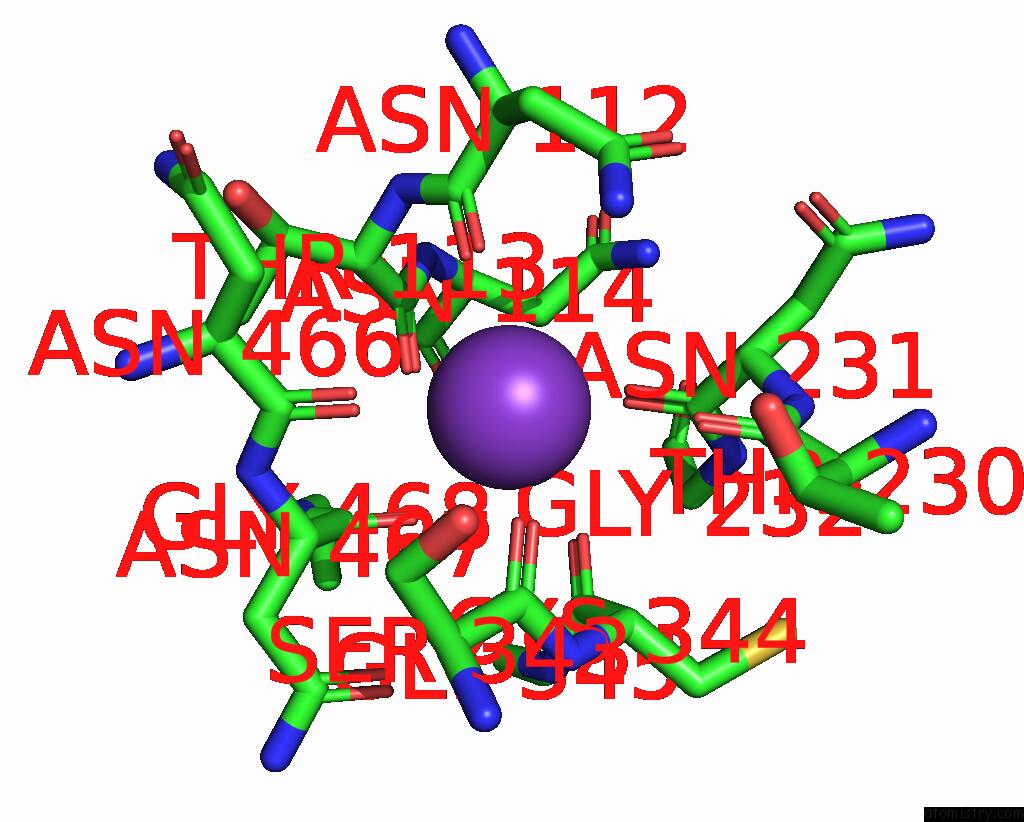
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 5 of Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+ within 5.0Å range:
|
Potassium binding site 6 out of 7 in 7zri
Go back to
Potassium binding site 6 out
of 7 in the Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+
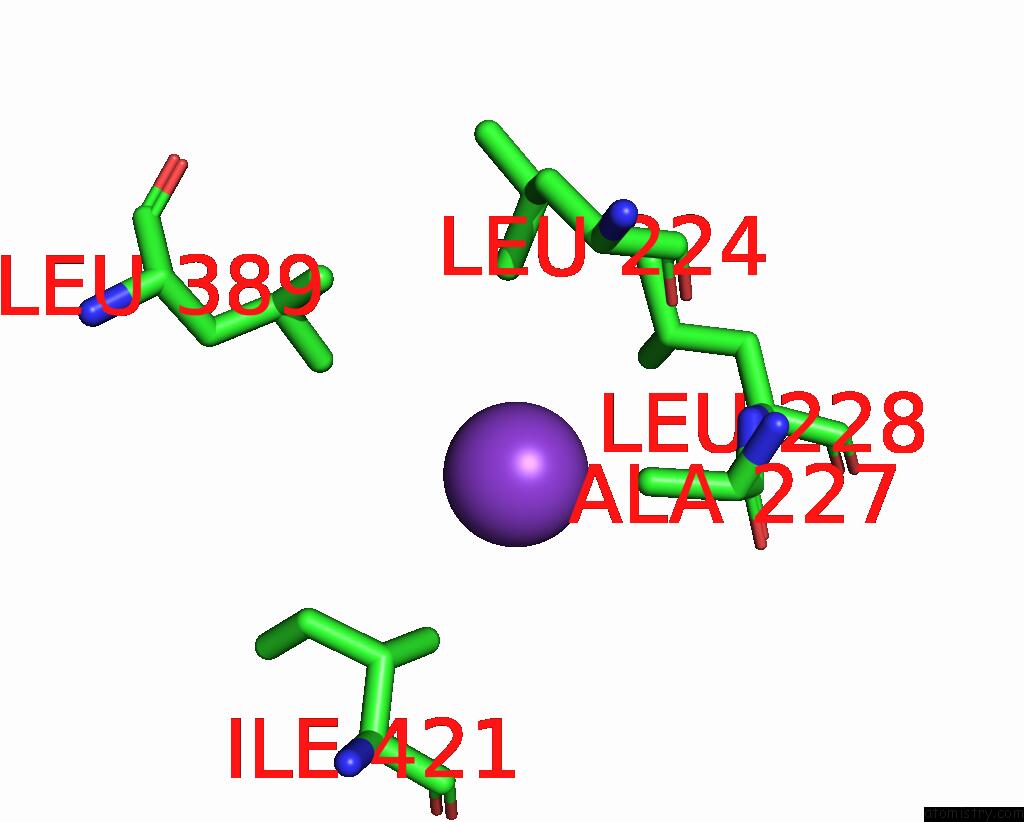
Mono view
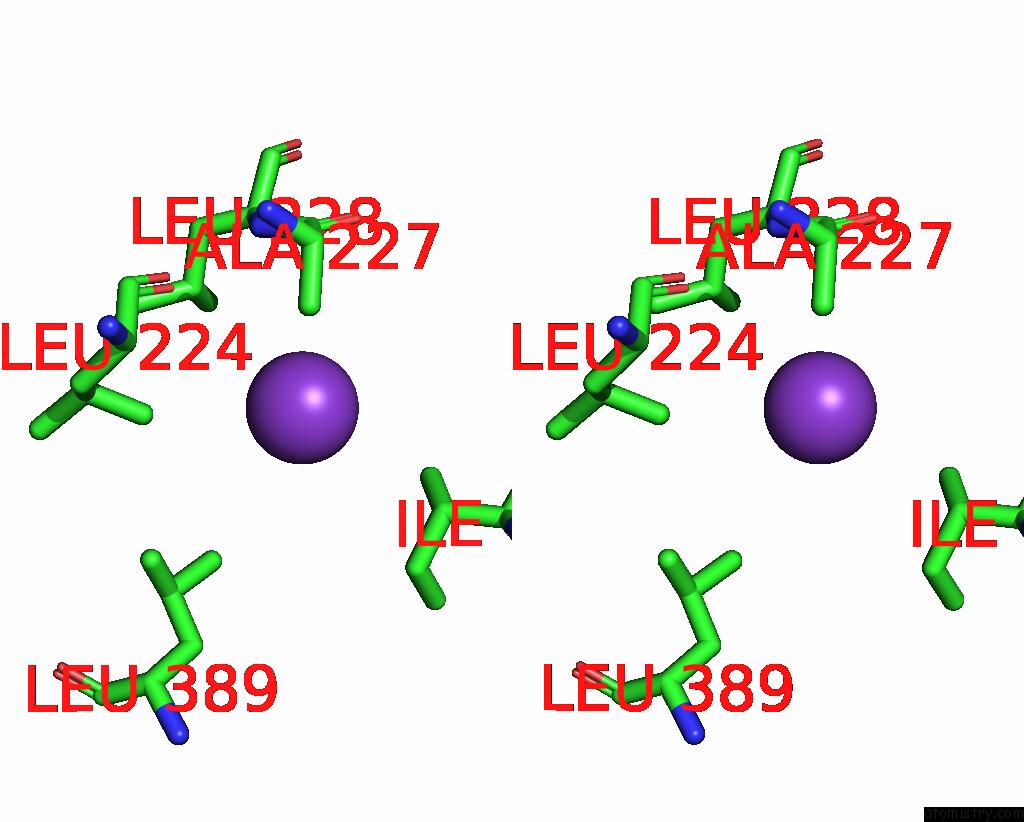
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 6 of Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+ within 5.0Å range:
|
Potassium binding site 7 out of 7 in 7zri
Go back to
Potassium binding site 7 out
of 7 in the Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+

Mono view
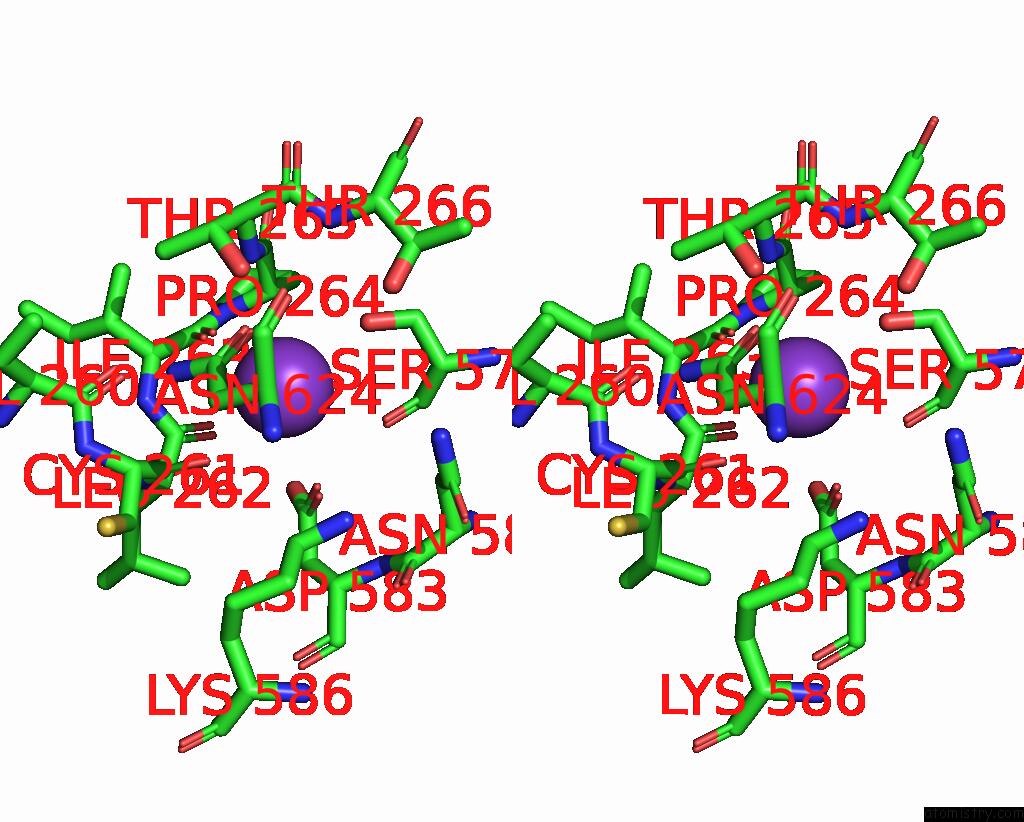
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 7 of Cryo-Em Structure of the Kdpfabc Complex in A Nucleotide-Free E1 Conformation Loaded with K+ within 5.0Å range:
|
Reference:
J.M.Silberberg,
C.Stock,
L.Hielkema,
R.A.Corey,
J.Rheinberger,
D.Wunnicke,
V.R.A.Dubach,
P.J.Stansfeld,
I.Hanelt,
C.Paulino.
Inhibited Kdpfabc Transitions Into An E1 Off-Cycle State. Elife V. 11 2022.
ISSN: ESSN 2050-084X
PubMed: 36255052
DOI: 10.7554/ELIFE.80988
Page generated: Mon Aug 12 21:56:09 2024
ISSN: ESSN 2050-084X
PubMed: 36255052
DOI: 10.7554/ELIFE.80988
Last articles
Zn in 9JPJZn in 9JP7
Zn in 9JPK
Zn in 9JPL
Zn in 9GN6
Zn in 9GN7
Zn in 9GKU
Zn in 9GKW
Zn in 9GKX
Zn in 9GL0