Potassium »
PDB 7tb0-7uui »
7tcs »
Potassium in PDB 7tcs: M379A Mutant Tyrosine Phenol-Lyase Complexed with L-Methionine
Enzymatic activity of M379A Mutant Tyrosine Phenol-Lyase Complexed with L-Methionine
All present enzymatic activity of M379A Mutant Tyrosine Phenol-Lyase Complexed with L-Methionine:
4.1.99.2;
4.1.99.2;
Protein crystallography data
The structure of M379A Mutant Tyrosine Phenol-Lyase Complexed with L-Methionine, PDB code: 7tcs
was solved by
R.S.Phillips,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.27 / 1.37 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 63.55, 82.99, 96.58, 93.73, 105.71, 114.24 |
| R / Rfree (%) | 14.1 / 15.5 |
Other elements in 7tcs:
The structure of M379A Mutant Tyrosine Phenol-Lyase Complexed with L-Methionine also contains other interesting chemical elements:
| Caesium | (Cs) | 3 atoms |
Potassium Binding Sites:
The binding sites of Potassium atom in the M379A Mutant Tyrosine Phenol-Lyase Complexed with L-Methionine
(pdb code 7tcs). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 4 binding sites of Potassium where determined in the M379A Mutant Tyrosine Phenol-Lyase Complexed with L-Methionine, PDB code: 7tcs:
Jump to Potassium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Potassium where determined in the M379A Mutant Tyrosine Phenol-Lyase Complexed with L-Methionine, PDB code: 7tcs:
Jump to Potassium binding site number: 1; 2; 3; 4;
Potassium binding site 1 out of 4 in 7tcs
Go back to
Potassium binding site 1 out
of 4 in the M379A Mutant Tyrosine Phenol-Lyase Complexed with L-Methionine

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of M379A Mutant Tyrosine Phenol-Lyase Complexed with L-Methionine within 5.0Å range:
|
Potassium binding site 2 out of 4 in 7tcs
Go back to
Potassium binding site 2 out
of 4 in the M379A Mutant Tyrosine Phenol-Lyase Complexed with L-Methionine

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of M379A Mutant Tyrosine Phenol-Lyase Complexed with L-Methionine within 5.0Å range:
|
Potassium binding site 3 out of 4 in 7tcs
Go back to
Potassium binding site 3 out
of 4 in the M379A Mutant Tyrosine Phenol-Lyase Complexed with L-Methionine

Mono view
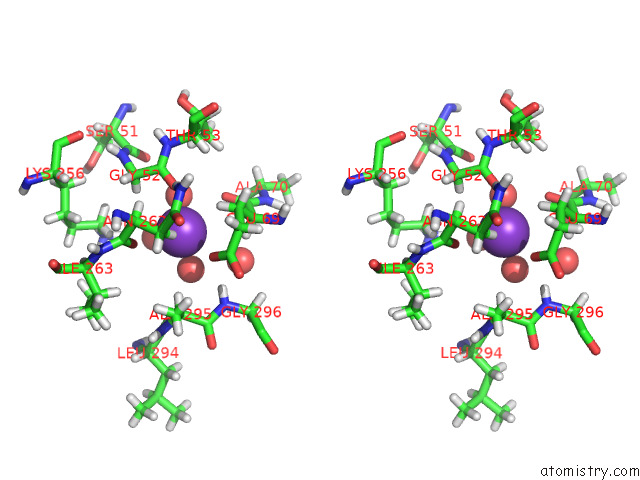
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 3 of M379A Mutant Tyrosine Phenol-Lyase Complexed with L-Methionine within 5.0Å range:
|
Potassium binding site 4 out of 4 in 7tcs
Go back to
Potassium binding site 4 out
of 4 in the M379A Mutant Tyrosine Phenol-Lyase Complexed with L-Methionine
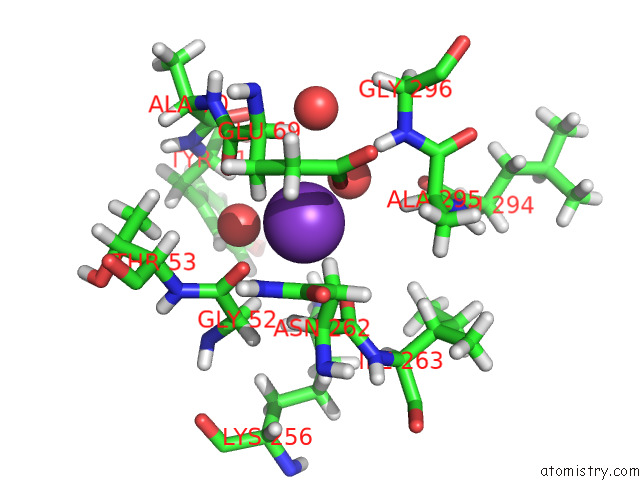
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 4 of M379A Mutant Tyrosine Phenol-Lyase Complexed with L-Methionine within 5.0Å range:
|
Reference:
R.S.Phillips,
B.Jones,
S.Nash.
M379A Mutant Tyrosine Phenol-Lyase From Citrobacter Freundii Has Altered Conformational Dynamics. Chembiochem V. 23 00028 2022.
ISSN: ESSN 1439-7633
PubMed: 35577764
DOI: 10.1002/CBIC.202200028
Page generated: Sat Aug 9 14:58:34 2025
ISSN: ESSN 1439-7633
PubMed: 35577764
DOI: 10.1002/CBIC.202200028
Last articles
Mg in 3JATMg in 3JAW
Mg in 3JAS
Mg in 3JAR
Mg in 3J81
Mg in 3JAO
Mg in 3JAL
Mg in 3JAK
Mg in 3JAA
Mg in 3J8I