Potassium »
PDB 7n9l-7ojz »
7nnl »
Potassium in PDB 7nnl: Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+
Enzymatic activity of Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+
All present enzymatic activity of Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+:
7.2.2.6;
7.2.2.6;
Potassium Binding Sites:
The binding sites of Potassium atom in the Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+
(pdb code 7nnl). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 10 binding sites of Potassium where determined in the Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+, PDB code: 7nnl:
Jump to Potassium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
In total 10 binding sites of Potassium where determined in the Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+, PDB code: 7nnl:
Jump to Potassium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Potassium binding site 1 out of 10 in 7nnl
Go back to
Potassium binding site 1 out
of 10 in the Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+
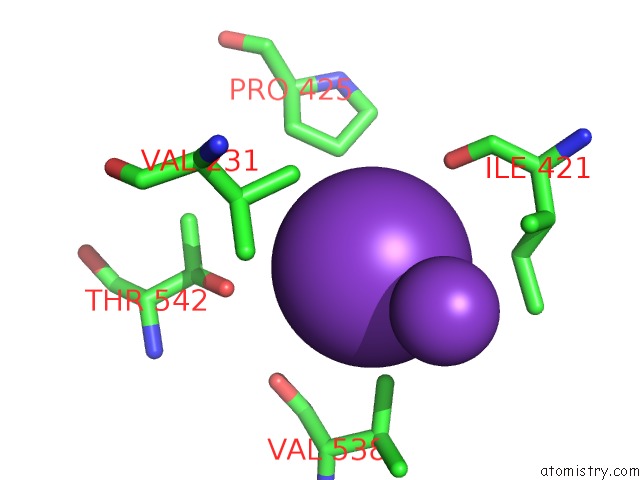
Mono view
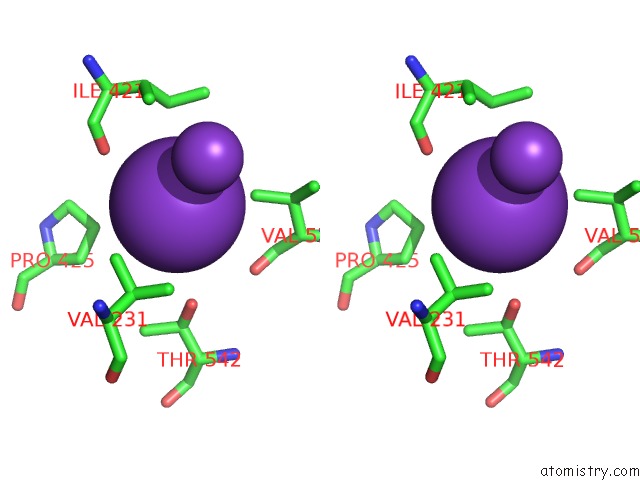
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+ within 5.0Å range:
|
Potassium binding site 2 out of 10 in 7nnl
Go back to
Potassium binding site 2 out
of 10 in the Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+
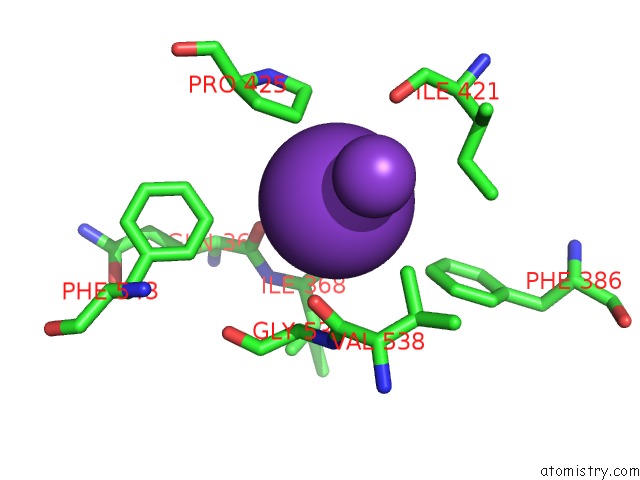
Mono view
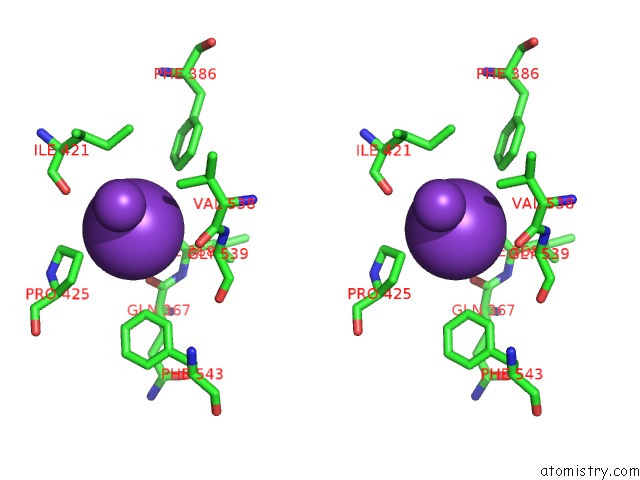
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+ within 5.0Å range:
|
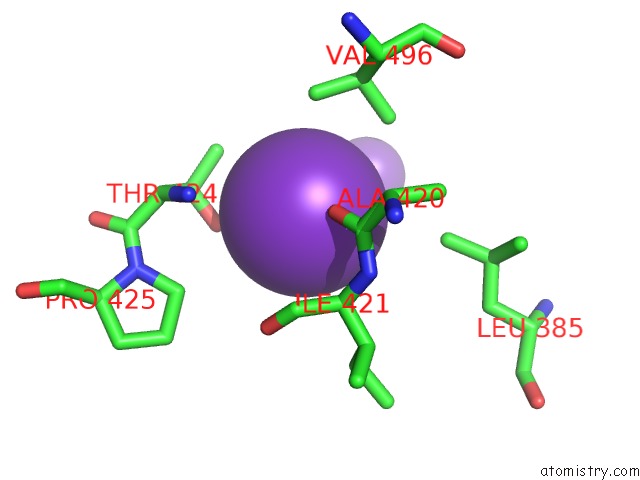
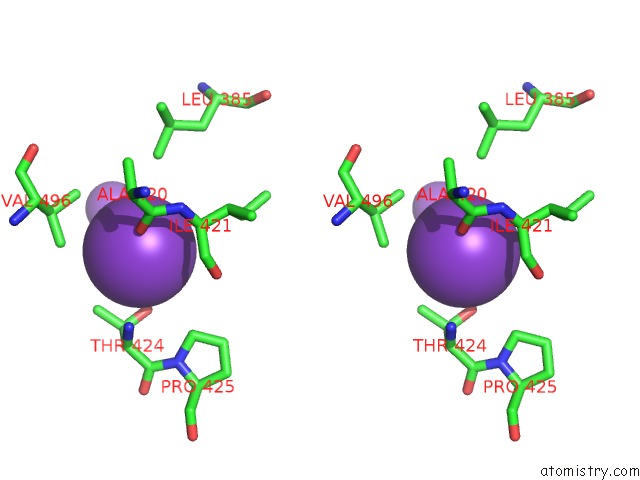
Potassium binding site 3 out of 10 in 7nnl
Go back to
Potassium binding site 3 out
of 10 in the Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 3 of Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+ within 5.0Å range:
|
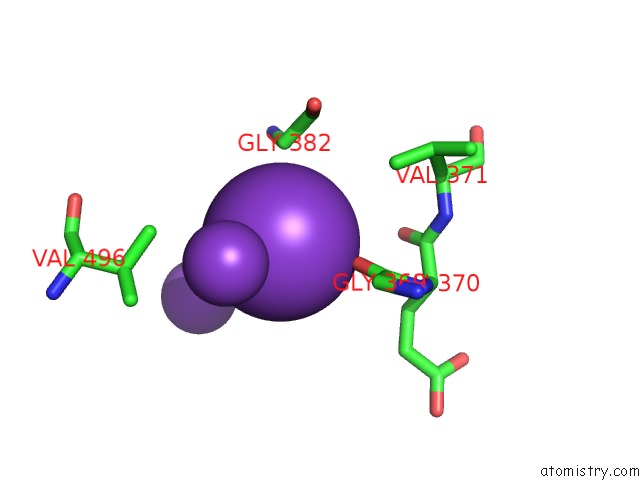
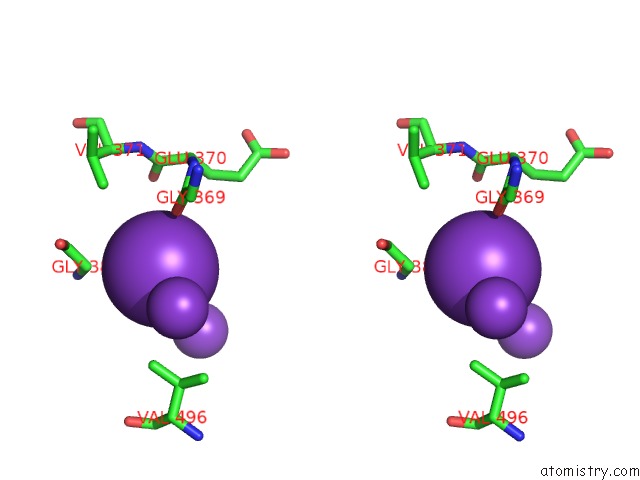
Potassium binding site 4 out of 10 in 7nnl
Go back to
Potassium binding site 4 out
of 10 in the Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 4 of Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+ within 5.0Å range:
|
Potassium binding site 5 out of 10 in 7nnl
Go back to
Potassium binding site 5 out
of 10 in the Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+
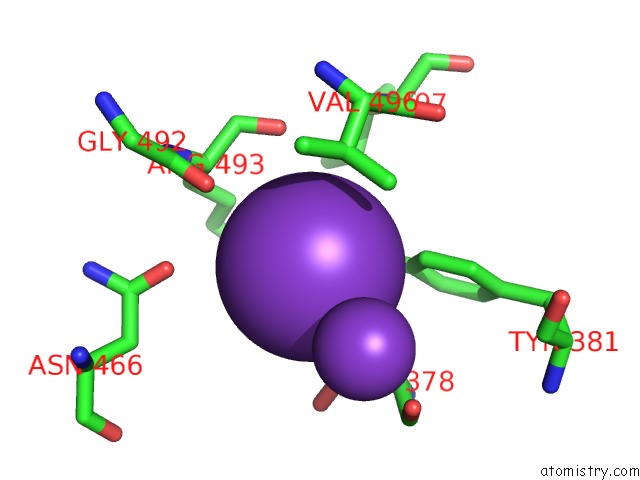
Mono view
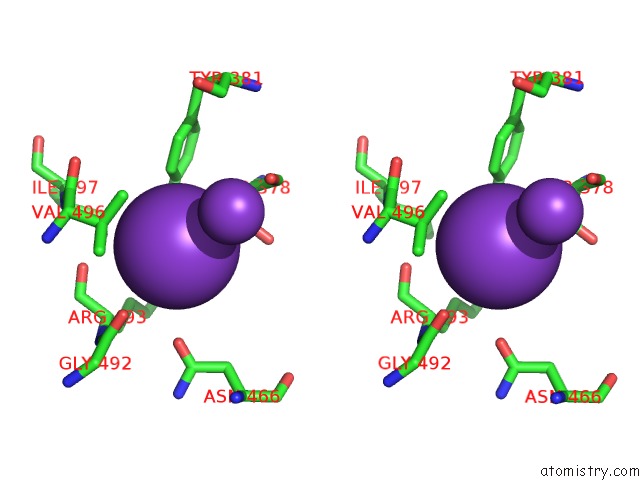
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 5 of Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+ within 5.0Å range:
|
Potassium binding site 6 out of 10 in 7nnl
Go back to
Potassium binding site 6 out
of 10 in the Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+
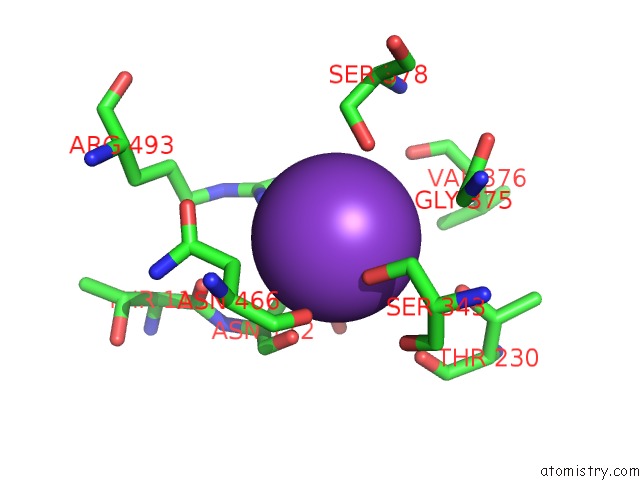
Mono view
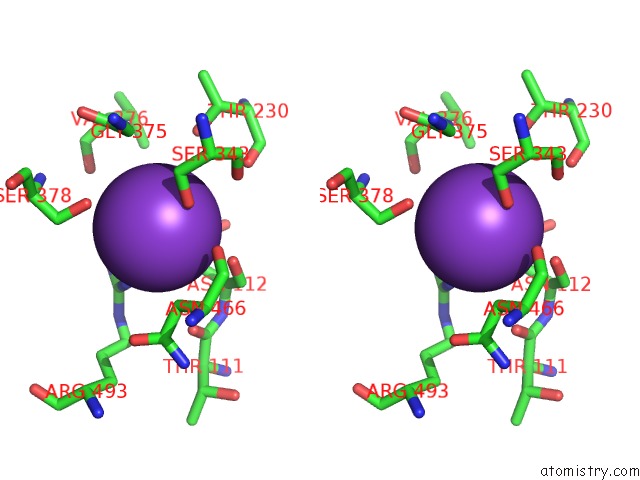
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 6 of Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+ within 5.0Å range:
|
Potassium binding site 7 out of 10 in 7nnl
Go back to
Potassium binding site 7 out
of 10 in the Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+
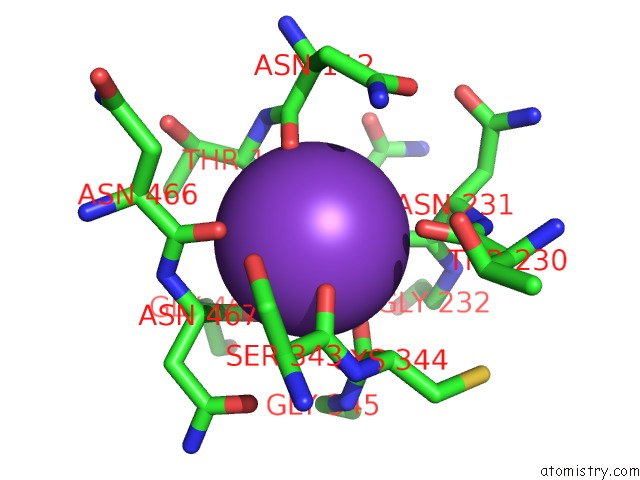
Mono view
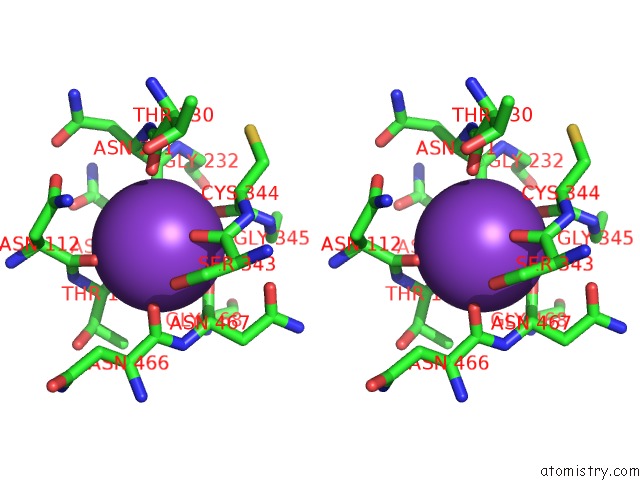
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 7 of Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+ within 5.0Å range:
|
Potassium binding site 8 out of 10 in 7nnl
Go back to
Potassium binding site 8 out
of 10 in the Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+
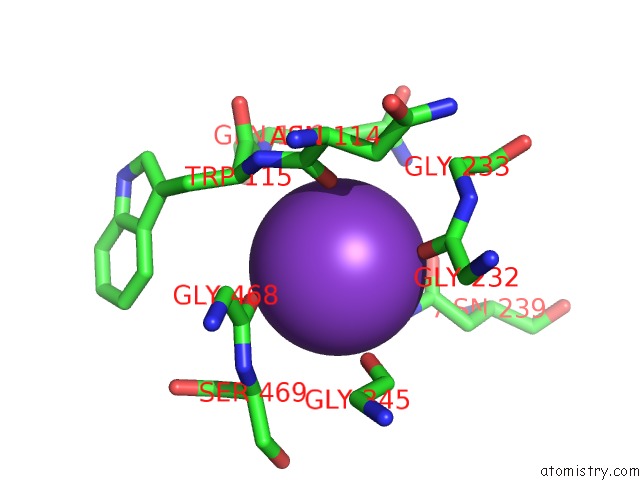
Mono view
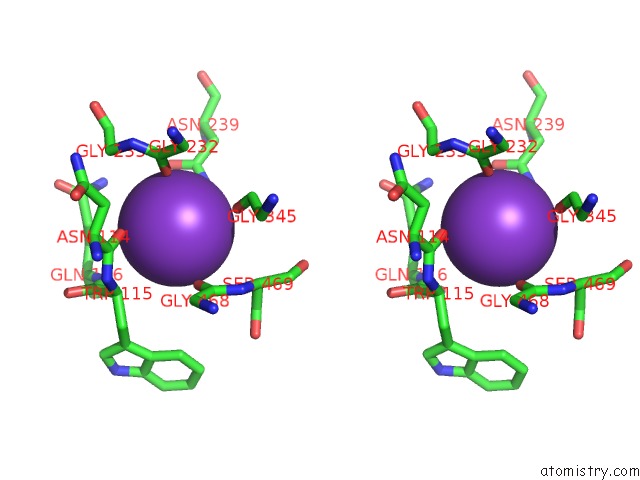
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 8 of Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+ within 5.0Å range:
|
Potassium binding site 9 out of 10 in 7nnl
Go back to
Potassium binding site 9 out
of 10 in the Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+
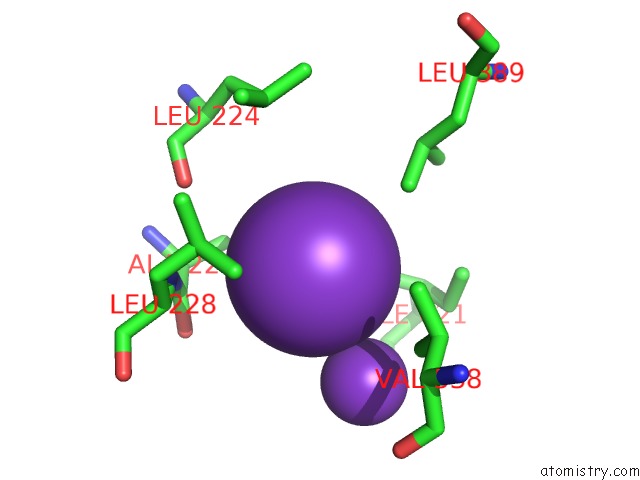
Mono view
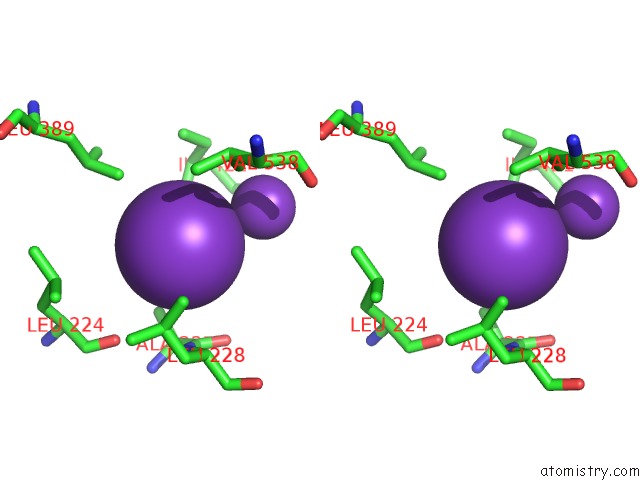
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 9 of Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+ within 5.0Å range:
|
Potassium binding site 10 out of 10 in 7nnl
Go back to
Potassium binding site 10 out
of 10 in the Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+
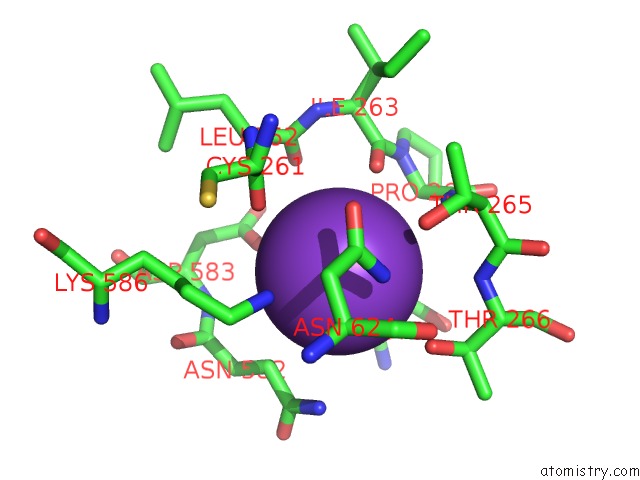
Mono view
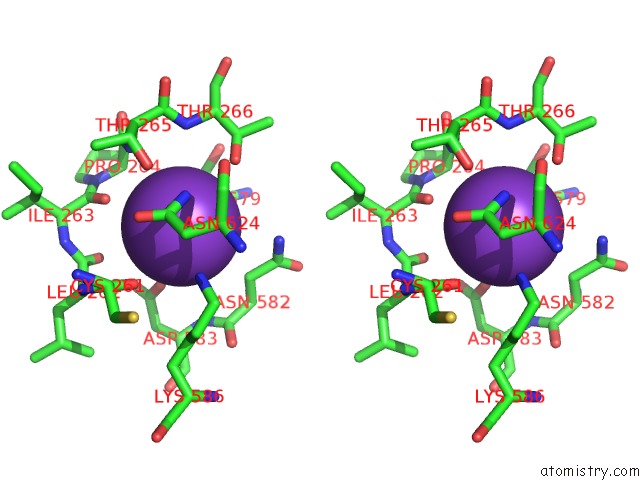
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 10 of Cryo-Em Structure of the Kdpfabc Complex in An E1-Atp Conformation Loaded with K+ within 5.0Å range:
|
Reference:
J.M.Silberberg,
R.A.Corey,
L.Hielkema,
C.Stock,
P.J.Stansfeld,
C.Paulino,
I.Hanelt.
Deciphering Ion Transport and Atpase Coupling in the Intersubunit Tunnel of Kdpfabc Biorxiv 2021.
DOI: 10.1101/2021.05.18.444724
Page generated: Sat Aug 9 13:45:11 2025
DOI: 10.1101/2021.05.18.444724
Last articles
Mg in 2IYNMg in 2IXF
Mg in 2IYW
Mg in 2IYF
Mg in 2IW4
Mg in 2IY5
Mg in 2IXE
Mg in 2IX1
Mg in 2IW9
Mg in 2IW6