Potassium »
PDB 6qml-6rv4 »
6rtg »
Potassium in PDB 6rtg: Crystal Structure of the Udp-Bound Glycosyltransferase Domain From the Ygt Toxin
Protein crystallography data
The structure of Crystal Structure of the Udp-Bound Glycosyltransferase Domain From the Ygt Toxin, PDB code: 6rtg
was solved by
C.Wirth,
X.Bogdanovic,
W.-C.Kao,
C.Hunte,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.56 / 1.90 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 164.270, 164.270, 47.740, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.6 / 22.3 |
Other elements in 6rtg:
The structure of Crystal Structure of the Udp-Bound Glycosyltransferase Domain From the Ygt Toxin also contains other interesting chemical elements:
| Manganese | (Mn) | 1 atom |
Potassium Binding Sites:
The binding sites of Potassium atom in the Crystal Structure of the Udp-Bound Glycosyltransferase Domain From the Ygt Toxin
(pdb code 6rtg). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total only one binding site of Potassium was determined in the Crystal Structure of the Udp-Bound Glycosyltransferase Domain From the Ygt Toxin, PDB code: 6rtg:
In total only one binding site of Potassium was determined in the Crystal Structure of the Udp-Bound Glycosyltransferase Domain From the Ygt Toxin, PDB code: 6rtg:
Potassium binding site 1 out of 1 in 6rtg
Go back to
Potassium binding site 1 out
of 1 in the Crystal Structure of the Udp-Bound Glycosyltransferase Domain From the Ygt Toxin
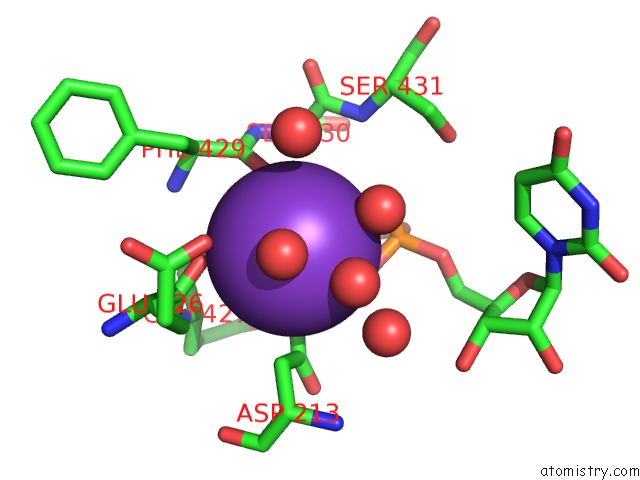
Mono view
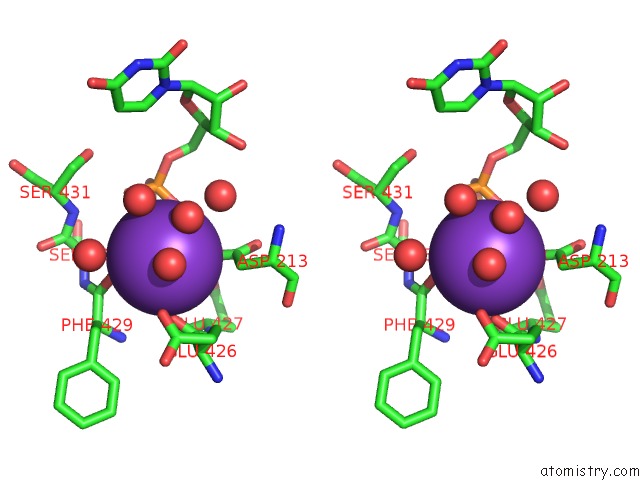
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Crystal Structure of the Udp-Bound Glycosyltransferase Domain From the Ygt Toxin within 5.0Å range:
|
Reference:
G.S.Ost,
C.Wirth,
X.Bogdanovic,
W.-C.Kao,
B.Schorch,
P.J.K.Aktories,
M.Westphal,
P.Papatheodorou,
C.Schwan,
A.Schlosser,
W.Driever,
T.Jank,
C.Hunte,
K.Aktories.
Inverse Control of Rab Proteins By Yersinia Adp-Ribosyltransferase and Glycosyltransferase Related to Clostridial Glucosylating Toxins Sci Adv 2020.
ISSN: ESSN 2375-2548
DOI: 10.1126/SCIADV.AAZ2094
Page generated: Sat Aug 9 12:04:08 2025
ISSN: ESSN 2375-2548
DOI: 10.1126/SCIADV.AAZ2094
Last articles
Mg in 2AI3Mg in 2AI2
Mg in 2AGP
Mg in 2AI1
Mg in 2AHO
Mg in 2AGQ
Mg in 2AGO
Mg in 2AFI
Mg in 2AG1
Mg in 2AG0