Potassium »
PDB 6qml-6rv4 »
6r32 »
Potassium in PDB 6r32: Aspergillus Niger Ferrulic Acid Decarboxylase (Fdc) in Complex with the Covalent Adduct Formed Between Prfmn Cofactor and Phenylpropiolic Acid (INT1')
Enzymatic activity of Aspergillus Niger Ferrulic Acid Decarboxylase (Fdc) in Complex with the Covalent Adduct Formed Between Prfmn Cofactor and Phenylpropiolic Acid (INT1')
All present enzymatic activity of Aspergillus Niger Ferrulic Acid Decarboxylase (Fdc) in Complex with the Covalent Adduct Formed Between Prfmn Cofactor and Phenylpropiolic Acid (INT1'):
4.1.1.102;
4.1.1.102;
Protein crystallography data
The structure of Aspergillus Niger Ferrulic Acid Decarboxylase (Fdc) in Complex with the Covalent Adduct Formed Between Prfmn Cofactor and Phenylpropiolic Acid (INT1'), PDB code: 6r32
was solved by
S.S.Bailey,
D.Leys,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 64.70 / 1.21 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 95.808, 64.265, 87.726, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 13 / 15.9 |
Other elements in 6r32:
The structure of Aspergillus Niger Ferrulic Acid Decarboxylase (Fdc) in Complex with the Covalent Adduct Formed Between Prfmn Cofactor and Phenylpropiolic Acid (INT1') also contains other interesting chemical elements:
| Manganese | (Mn) | 1 atom |
Potassium Binding Sites:
The binding sites of Potassium atom in the Aspergillus Niger Ferrulic Acid Decarboxylase (Fdc) in Complex with the Covalent Adduct Formed Between Prfmn Cofactor and Phenylpropiolic Acid (INT1')
(pdb code 6r32). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 2 binding sites of Potassium where determined in the Aspergillus Niger Ferrulic Acid Decarboxylase (Fdc) in Complex with the Covalent Adduct Formed Between Prfmn Cofactor and Phenylpropiolic Acid (INT1'), PDB code: 6r32:
Jump to Potassium binding site number: 1; 2;
In total 2 binding sites of Potassium where determined in the Aspergillus Niger Ferrulic Acid Decarboxylase (Fdc) in Complex with the Covalent Adduct Formed Between Prfmn Cofactor and Phenylpropiolic Acid (INT1'), PDB code: 6r32:
Jump to Potassium binding site number: 1; 2;
Potassium binding site 1 out of 2 in 6r32
Go back to
Potassium binding site 1 out
of 2 in the Aspergillus Niger Ferrulic Acid Decarboxylase (Fdc) in Complex with the Covalent Adduct Formed Between Prfmn Cofactor and Phenylpropiolic Acid (INT1')
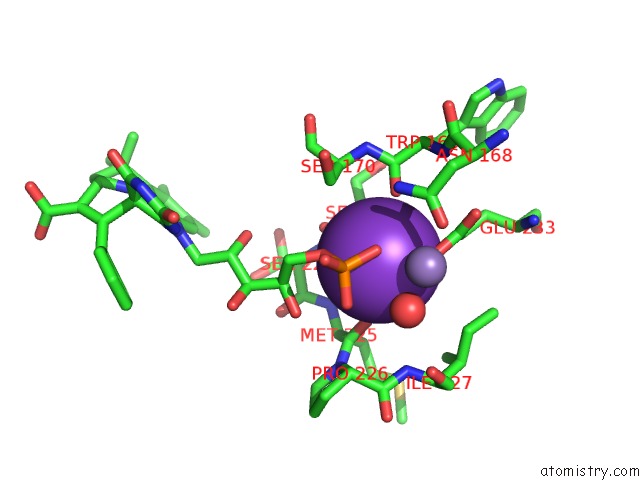
Mono view
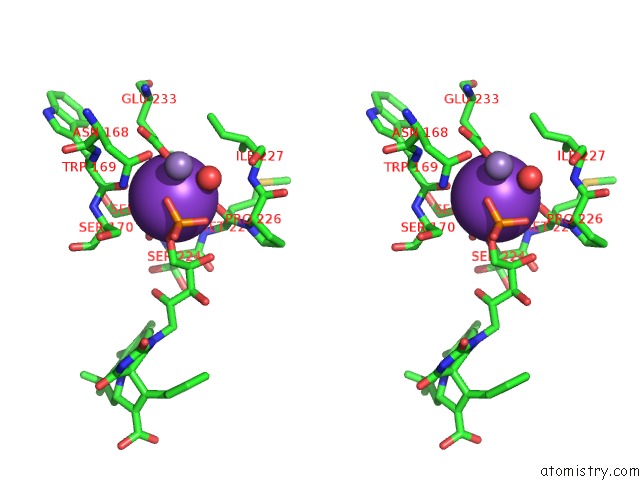
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Aspergillus Niger Ferrulic Acid Decarboxylase (Fdc) in Complex with the Covalent Adduct Formed Between Prfmn Cofactor and Phenylpropiolic Acid (INT1') within 5.0Å range:
|
Potassium binding site 2 out of 2 in 6r32
Go back to
Potassium binding site 2 out
of 2 in the Aspergillus Niger Ferrulic Acid Decarboxylase (Fdc) in Complex with the Covalent Adduct Formed Between Prfmn Cofactor and Phenylpropiolic Acid (INT1')
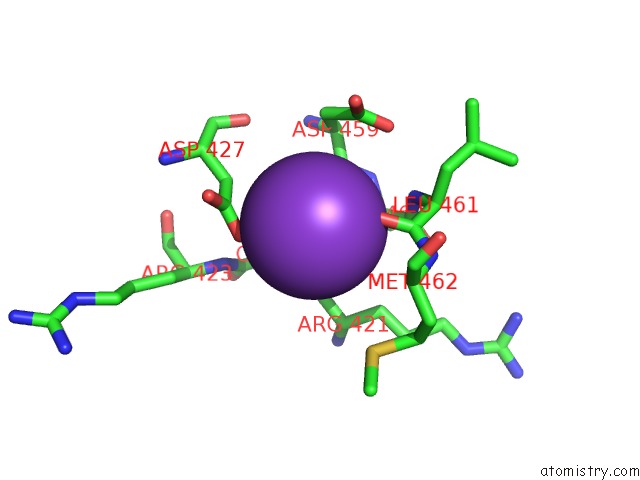
Mono view
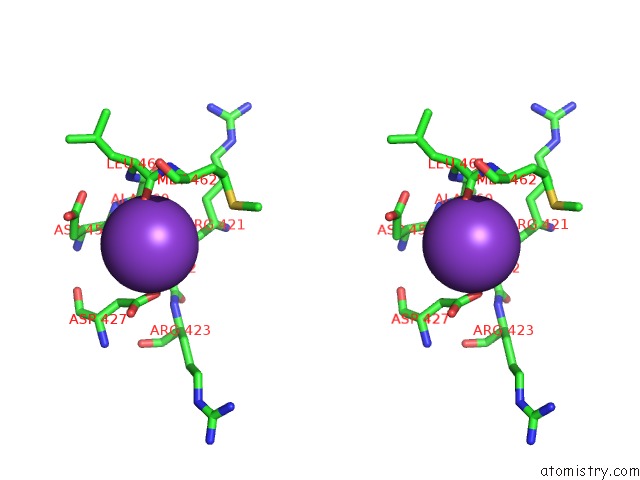
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Aspergillus Niger Ferrulic Acid Decarboxylase (Fdc) in Complex with the Covalent Adduct Formed Between Prfmn Cofactor and Phenylpropiolic Acid (INT1') within 5.0Å range:
|
Reference:
S.S.Bailey,
K.A.P.Payne,
A.Saaret,
S.A.Marshall,
I.Gostimskaya,
I.Kosov,
K.Fisher,
S.Hay,
D.Leys.
Atomic Description of An Enzyme Reaction Dependent on Reversible 1,3-Dipolar Cycloaddition To Be Published.
Page generated: Sat Aug 9 12:00:16 2025
Last articles
Mg in 1YLSMg in 1YL7
Mg in 1YL6
Mg in 1YL5
Mg in 1YKV
Mg in 1YKQ
Mg in 1YIT
Mg in 1YIJ
Mg in 1YJF
Mg in 1YI2