Potassium »
PDB 5sc8-5u3q »
5tkr »
Potassium in PDB 5tkr: Crystal Structure of A Lipomyces Starkeyi Levoglucosan Kinase G359R Mutant
Protein crystallography data
The structure of Crystal Structure of A Lipomyces Starkeyi Levoglucosan Kinase G359R Mutant, PDB code: 5tkr
was solved by
J.P.Bacik,
J.R.Klesmith,
R.Michalczyk,
T.A.Whitehead,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.08 / 1.80 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 70.060, 70.060, 261.770, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.5 / 20.8 |
Other elements in 5tkr:
The structure of Crystal Structure of A Lipomyces Starkeyi Levoglucosan Kinase G359R Mutant also contains other interesting chemical elements:
| Magnesium | (Mg) | 2 atoms |
Potassium Binding Sites:
The binding sites of Potassium atom in the Crystal Structure of A Lipomyces Starkeyi Levoglucosan Kinase G359R Mutant
(pdb code 5tkr). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 2 binding sites of Potassium where determined in the Crystal Structure of A Lipomyces Starkeyi Levoglucosan Kinase G359R Mutant, PDB code: 5tkr:
Jump to Potassium binding site number: 1; 2;
In total 2 binding sites of Potassium where determined in the Crystal Structure of A Lipomyces Starkeyi Levoglucosan Kinase G359R Mutant, PDB code: 5tkr:
Jump to Potassium binding site number: 1; 2;
Potassium binding site 1 out of 2 in 5tkr
Go back to
Potassium binding site 1 out
of 2 in the Crystal Structure of A Lipomyces Starkeyi Levoglucosan Kinase G359R Mutant
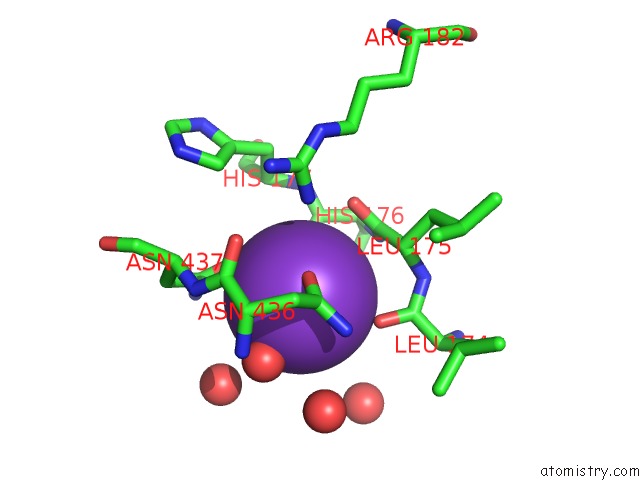
Mono view
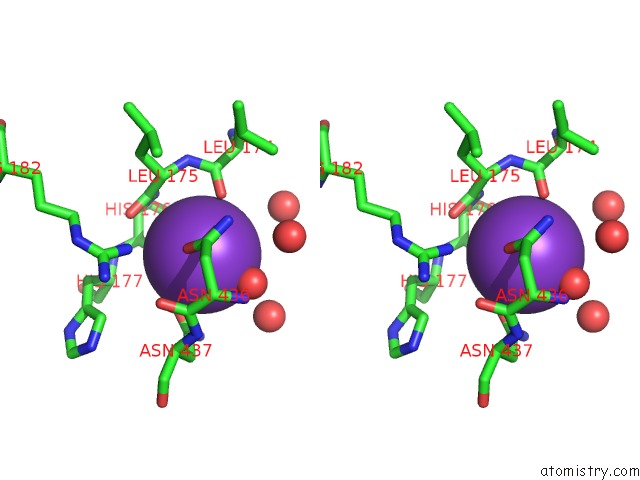
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Crystal Structure of A Lipomyces Starkeyi Levoglucosan Kinase G359R Mutant within 5.0Å range:
|
Potassium binding site 2 out of 2 in 5tkr
Go back to
Potassium binding site 2 out
of 2 in the Crystal Structure of A Lipomyces Starkeyi Levoglucosan Kinase G359R Mutant
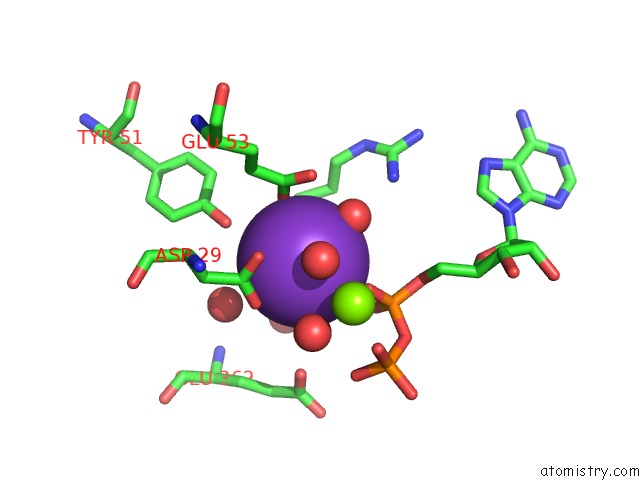
Mono view
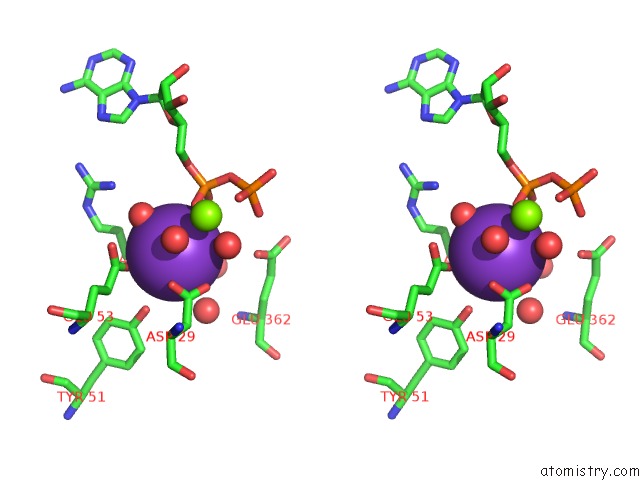
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Crystal Structure of A Lipomyces Starkeyi Levoglucosan Kinase G359R Mutant within 5.0Å range:
|
Reference:
J.R.Klesmith,
J.P.Bacik,
E.E.Wrenbeck,
R.Michalczyk,
T.A.Whitehead.
Trade-Offs Between Enzyme Fitness and Solubility Illuminated By Deep Mutational Scanning. Proc. Natl. Acad. Sci. V. 114 2265 2017U.S.A..
ISSN: ESSN 1091-6490
PubMed: 28196882
DOI: 10.1073/PNAS.1614437114
Page generated: Sat Aug 9 09:50:18 2025
ISSN: ESSN 1091-6490
PubMed: 28196882
DOI: 10.1073/PNAS.1614437114
Last articles
Mg in 1N67Mg in 1N6I
Mg in 1N6H
Mg in 1N5Y
Mg in 1N5L
Mg in 1N5K
Mg in 1N5J
Mg in 1N56
Mg in 1N57
Mg in 1N52