Potassium »
PDB 3ukm-3vrs »
3vks »
Potassium in PDB 3vks: Assimilatory Nitrite Reductase (NII3) - No Complex From Tobbaco Leaf
Enzymatic activity of Assimilatory Nitrite Reductase (NII3) - No Complex From Tobbaco Leaf
All present enzymatic activity of Assimilatory Nitrite Reductase (NII3) - No Complex From Tobbaco Leaf:
1.7.7.1;
1.7.7.1;
Protein crystallography data
The structure of Assimilatory Nitrite Reductase (NII3) - No Complex From Tobbaco Leaf, PDB code: 3vks
was solved by
S.Nakano,
M.Takahashi,
A.Sakamoto,
H.Morikawa,
K.Katayanagi,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 23.49 / 1.40 |
| Space group | P 4 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 132.758, 132.758, 77.539, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.1 / 16.8 |
Other elements in 3vks:
The structure of Assimilatory Nitrite Reductase (NII3) - No Complex From Tobbaco Leaf also contains other interesting chemical elements:
| Iron | (Fe) | 5 atoms |
| Chlorine | (Cl) | 2 atoms |
Potassium Binding Sites:
The binding sites of Potassium atom in the Assimilatory Nitrite Reductase (NII3) - No Complex From Tobbaco Leaf
(pdb code 3vks). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total only one binding site of Potassium was determined in the Assimilatory Nitrite Reductase (NII3) - No Complex From Tobbaco Leaf, PDB code: 3vks:
In total only one binding site of Potassium was determined in the Assimilatory Nitrite Reductase (NII3) - No Complex From Tobbaco Leaf, PDB code: 3vks:
Potassium binding site 1 out of 1 in 3vks
Go back to
Potassium binding site 1 out
of 1 in the Assimilatory Nitrite Reductase (NII3) - No Complex From Tobbaco Leaf
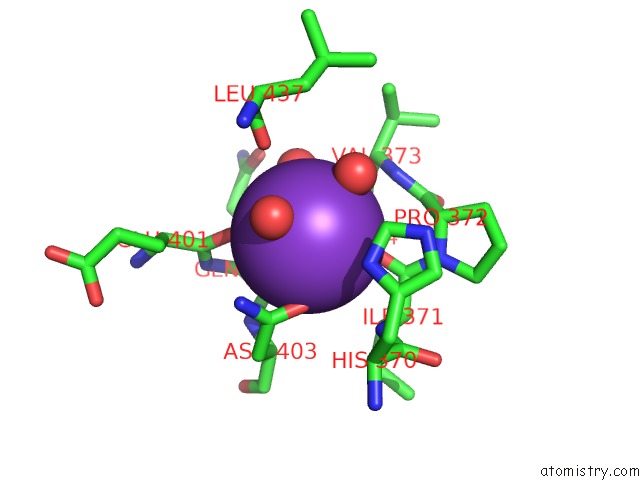
Mono view
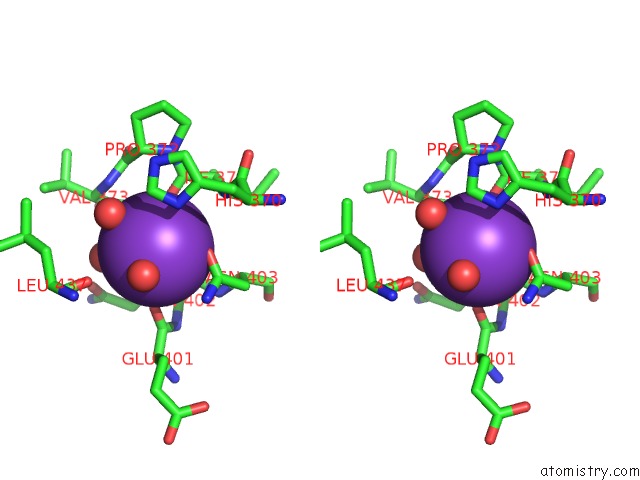
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Assimilatory Nitrite Reductase (NII3) - No Complex From Tobbaco Leaf within 5.0Å range:
|
Reference:
S.Nakano,
M.Takahashi,
A.Sakamoto,
H.Morikawa,
K.Katayanagi.
The Reductive Reaction Mechanism of Tobacco Nitrite Reductase Derived From A Combination of Crystal Structures and Ultraviolet-Visible Microspectroscopy Proteins V. 80 2035 2012.
ISSN: ISSN 0887-3585
PubMed: 22499059
DOI: 10.1002/PROT.24094
Page generated: Sat Aug 9 06:05:49 2025
ISSN: ISSN 0887-3585
PubMed: 22499059
DOI: 10.1002/PROT.24094
Last articles
K in 7LJ5K in 7LJ4
K in 7LFT
K in 7LC3
K in 7LFQ
K in 7LC6
K in 7L4G
K in 7L0Z
K in 7KZ0
K in 7L1A