Potassium »
PDB 1w2b-1yj9 »
1wxx »
Potassium in PDB 1wxx: Crystal Structure of TT1595, A Putative Sam-Dependent Methyltransferase From Thermus Thermophillus HB8
Protein crystallography data
The structure of Crystal Structure of TT1595, A Putative Sam-Dependent Methyltransferase From Thermus Thermophillus HB8, PDB code: 1wxx
was solved by
A.A.Pioszak,
K.Murayama,
N.Nakagawa,
A.Ebihara,
S.Kuramitsu,
M.Shirouzu,
S.Yokoyama,
Riken Structural Genomics/Proteomics Initiative (Rsgi),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 39.55 / 1.80 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 77.551, 82.318, 79.616, 96.34, 90.14, 103.06 |
| R / Rfree (%) | 20.3 / 22.9 |
Potassium Binding Sites:
The binding sites of Potassium atom in the Crystal Structure of TT1595, A Putative Sam-Dependent Methyltransferase From Thermus Thermophillus HB8
(pdb code 1wxx). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 4 binding sites of Potassium where determined in the Crystal Structure of TT1595, A Putative Sam-Dependent Methyltransferase From Thermus Thermophillus HB8, PDB code: 1wxx:
Jump to Potassium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Potassium where determined in the Crystal Structure of TT1595, A Putative Sam-Dependent Methyltransferase From Thermus Thermophillus HB8, PDB code: 1wxx:
Jump to Potassium binding site number: 1; 2; 3; 4;
Potassium binding site 1 out of 4 in 1wxx
Go back to
Potassium binding site 1 out
of 4 in the Crystal Structure of TT1595, A Putative Sam-Dependent Methyltransferase From Thermus Thermophillus HB8
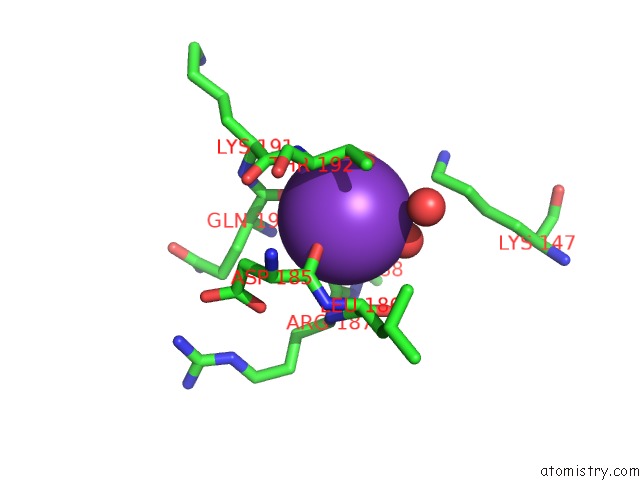
Mono view
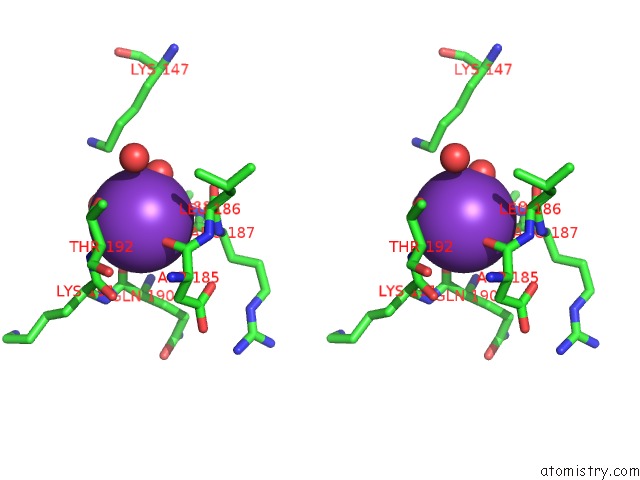
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Crystal Structure of TT1595, A Putative Sam-Dependent Methyltransferase From Thermus Thermophillus HB8 within 5.0Å range:
|
Potassium binding site 2 out of 4 in 1wxx
Go back to
Potassium binding site 2 out
of 4 in the Crystal Structure of TT1595, A Putative Sam-Dependent Methyltransferase From Thermus Thermophillus HB8
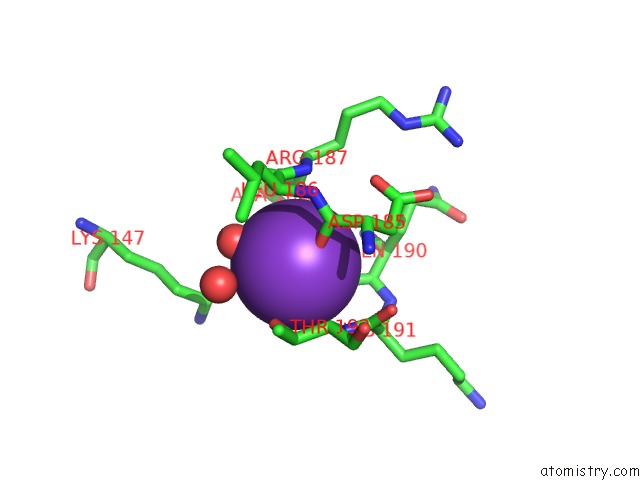
Mono view
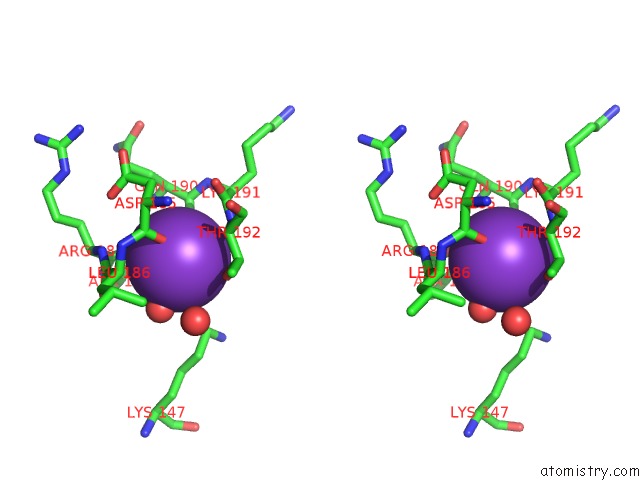
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Crystal Structure of TT1595, A Putative Sam-Dependent Methyltransferase From Thermus Thermophillus HB8 within 5.0Å range:
|
Potassium binding site 3 out of 4 in 1wxx
Go back to
Potassium binding site 3 out
of 4 in the Crystal Structure of TT1595, A Putative Sam-Dependent Methyltransferase From Thermus Thermophillus HB8
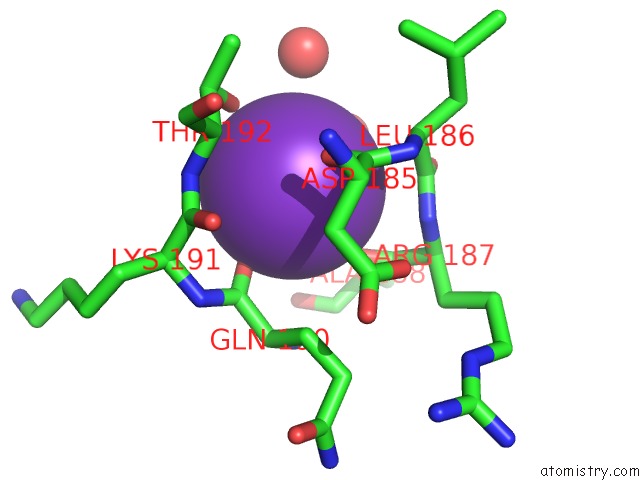
Mono view
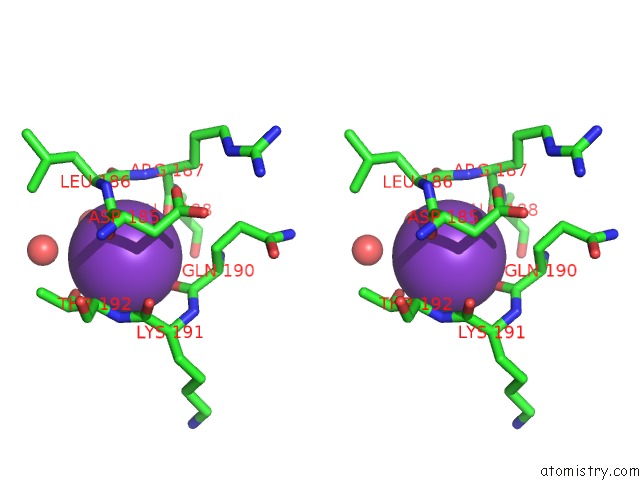
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 3 of Crystal Structure of TT1595, A Putative Sam-Dependent Methyltransferase From Thermus Thermophillus HB8 within 5.0Å range:
|
Potassium binding site 4 out of 4 in 1wxx
Go back to
Potassium binding site 4 out
of 4 in the Crystal Structure of TT1595, A Putative Sam-Dependent Methyltransferase From Thermus Thermophillus HB8
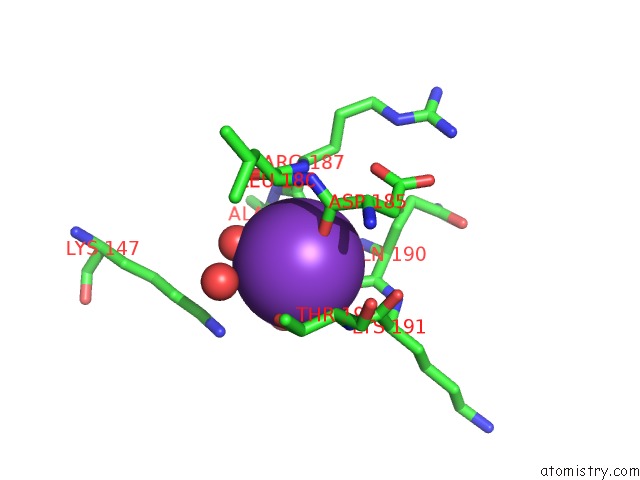
Mono view
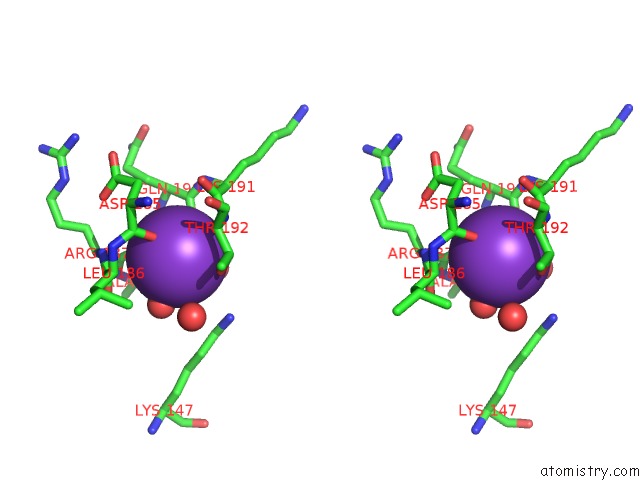
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 4 of Crystal Structure of TT1595, A Putative Sam-Dependent Methyltransferase From Thermus Thermophillus HB8 within 5.0Å range:
|
Reference:
A.A.Pioszak,
K.Murayama,
N.Nakagawa,
A.Ebihara,
S.Kuramitsu,
M.Shirouzu,
S.Yokoyama.
Structures of A Putative Rna 5-Methyluridine Methyltransferase, Thermus Thermophilus TTHA1280, and Its Complex with S-Adenosyl-L-Homocysteine. Acta Crystallogr.,Sect.F V. 61 867 2005.
ISSN: ESSN 1744-3091
PubMed: 16511182
DOI: 10.1107/S1744309105029842
Page generated: Sat Aug 9 02:53:13 2025
ISSN: ESSN 1744-3091
PubMed: 16511182
DOI: 10.1107/S1744309105029842
Last articles
K in 6DE8K in 6D1P
K in 6D2J
K in 6CW8
K in 6CSS
K in 6CST
K in 6CZ3
K in 6CSR
K in 6CSQ
K in 6CAO