Potassium in PDB 8w9d: Cryo-Em Structure of the RPD3S-Nucleosome Complex From Budding Yeast in State 1
Other elements in 8w9d:
The structure of Cryo-Em Structure of the RPD3S-Nucleosome Complex From Budding Yeast in State 1 also contains other interesting chemical elements:
| Zinc | (Zn) | 7 atoms |
Potassium Binding Sites:
The binding sites of Potassium atom in the Cryo-Em Structure of the RPD3S-Nucleosome Complex From Budding Yeast in State 1
(pdb code 8w9d). This binding sites where shown within
5.0 Angstroms radius around Potassium atom.
In total 2 binding sites of Potassium where determined in the Cryo-Em Structure of the RPD3S-Nucleosome Complex From Budding Yeast in State 1, PDB code: 8w9d:
Jump to Potassium binding site number: 1; 2;
In total 2 binding sites of Potassium where determined in the Cryo-Em Structure of the RPD3S-Nucleosome Complex From Budding Yeast in State 1, PDB code: 8w9d:
Jump to Potassium binding site number: 1; 2;
Potassium binding site 1 out of 2 in 8w9d
Go back to
Potassium binding site 1 out
of 2 in the Cryo-Em Structure of the RPD3S-Nucleosome Complex From Budding Yeast in State 1
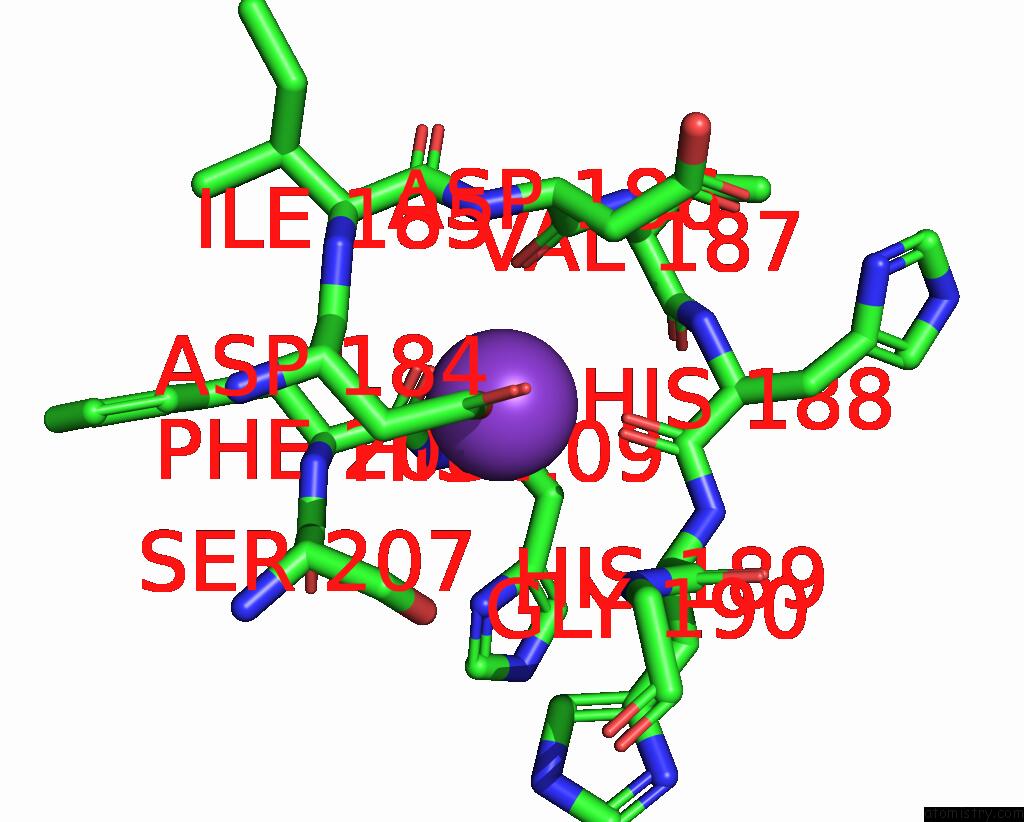
Mono view
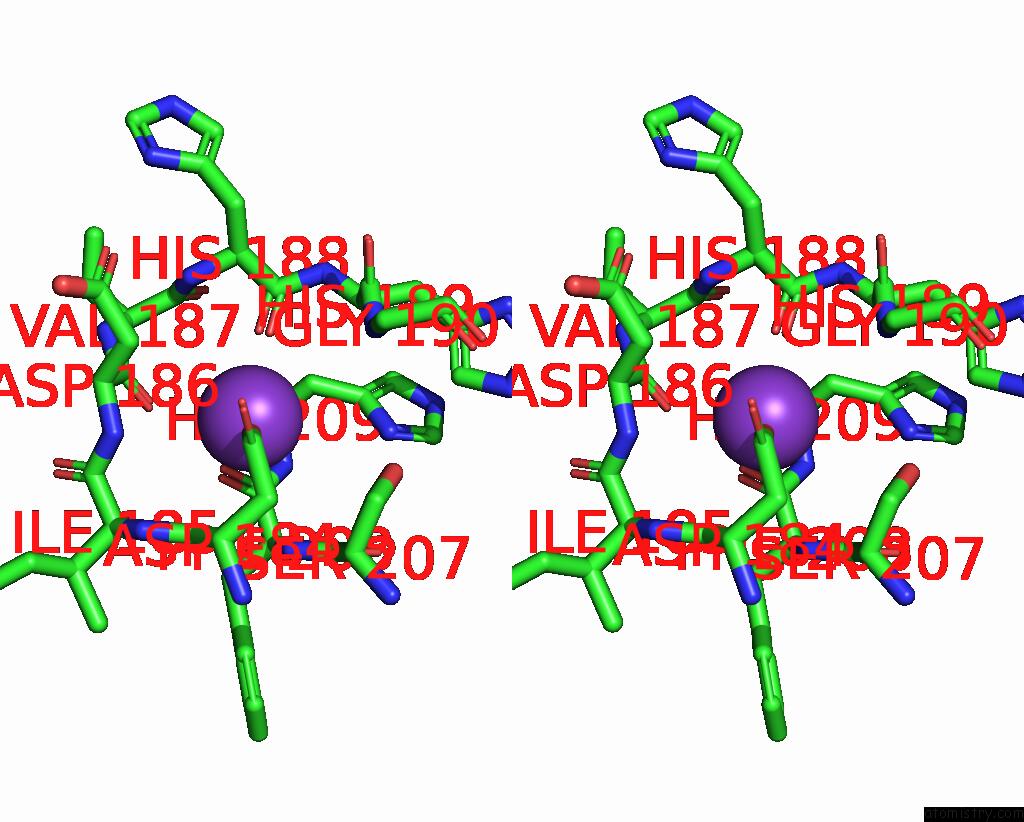
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 1 of Cryo-Em Structure of the RPD3S-Nucleosome Complex From Budding Yeast in State 1 within 5.0Å range:
|
Potassium binding site 2 out of 2 in 8w9d
Go back to
Potassium binding site 2 out
of 2 in the Cryo-Em Structure of the RPD3S-Nucleosome Complex From Budding Yeast in State 1
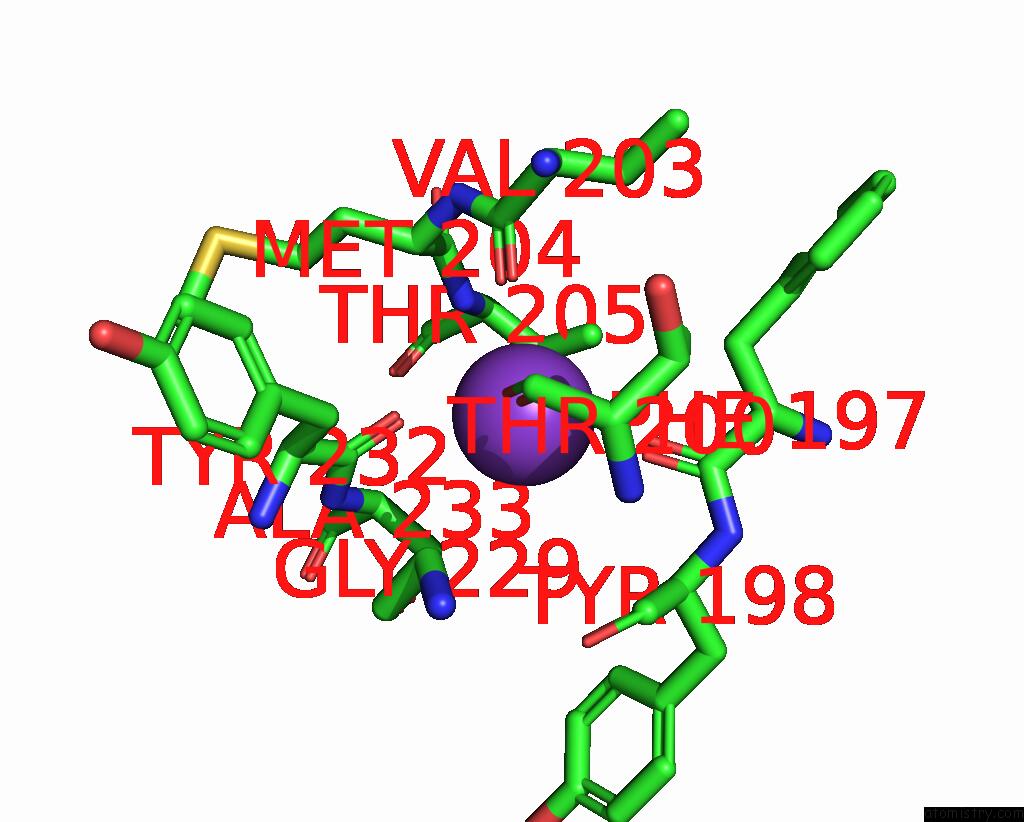
Mono view
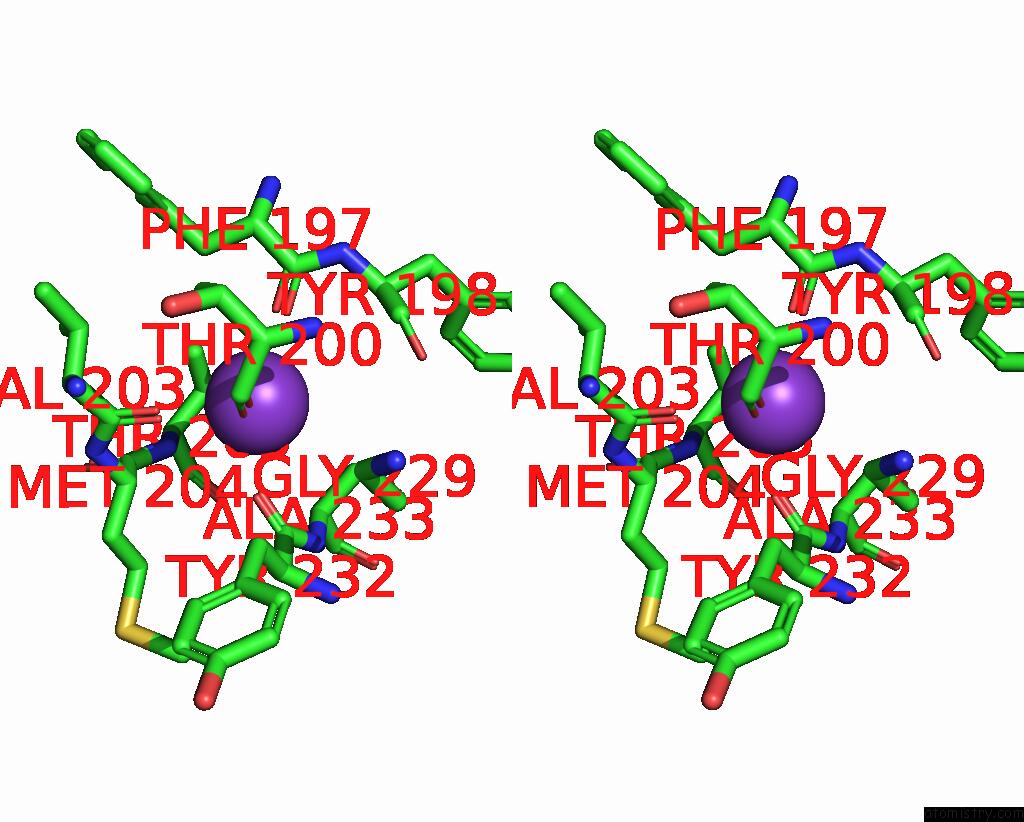
Stereo pair view

Mono view

Stereo pair view
A full contact list of Potassium with other atoms in the K binding
site number 2 of Cryo-Em Structure of the RPD3S-Nucleosome Complex From Budding Yeast in State 1 within 5.0Å range:
|
Reference:
C.Wang,
C.Chu,
Z.Guo,
X.Zhan.
Structures and Dynamics of RPD3S Complex Bound to Nucleosome. Sci Adv V. 10 K7678 2024.
ISSN: ESSN 2375-2548
PubMed: 38598631
DOI: 10.1126/SCIADV.ADK7678
Page generated: Tue Aug 13 01:13:24 2024
ISSN: ESSN 2375-2548
PubMed: 38598631
DOI: 10.1126/SCIADV.ADK7678
Last articles
Zn in 9IRQZn in 9IYX
Zn in 9J8P
Zn in 9IUU
Zn in 9GBF
Zn in 9G2V
Zn in 9G2L
Zn in 9G2X
Zn in 9G2Z
Zn in 9G2K